Human evolution: testing the molecular basis Understand article
In the second of two articles, Jarek Bryk describes how scientists dig deep into our genes – to test the molecular basis of an evolutionary adaptation in humans.

iStockphoto
The DNA of every organism holds information about its recent and ancient evolutionary history. By studying the patterns and changes in the DNA sequence – comparing the sequences between different individuals or species – we can uncover what has happened to them. We can find out which genes or fragments of the genome are likely to have provided an advantage to those individuals and species that carried them, allowing for their better survival and reproduction (see glossary for all terms in bold).
In a previous article (Bryk, 2010), I described a few examples of such beneficial genetic changes in humans and other organisms. Demonstrating which genetic changes might have been beneficial is difficult – especially in humans – but an even greater challenge is demonstrating the mechanism by which these changes could have improved the organisms’ survival and reproduction.
In this article, I present one of the approaches that scientists can use first to identify regions of our genome that could have helped us survive and reproduce, and then to test how these regions might have provided our ancestors with an advantage.
One of the ways potentially beneficial regions of our genome can be identified is simply by comparing DNA sequences of many individuals from different populations. In a very simple scenario, if one of these populations has been under selective pressure (for example, high UV radiation in a sunny region) that was absent in the other populations, the DNA sequence responsible for an appropriate adaptation (for example, a darker skin colour) should be different.
Adapted by with permission from Macmillan Publishers Ltd: Nature, Sabeti et al. (2007), © 2007
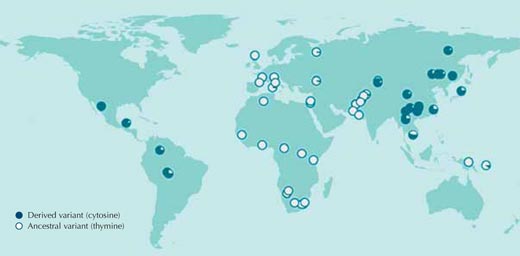
In the vast majority of cases, however, we do not know what selective pressures populations were exposed to in the past, or which genetic sequences are responsible for the adaptations. Let us begin, therefore, by comparing DNA sequences between human populations without any assumptions about what we may find. Figure 1 shows one such comparison, for a single nucleotide of the human genome.
When individuals have different nucleotides at a particular position in the DNA sequence, we call this a single nucleotide polymorphism (SNP, pronounced ‘snip’); three million such variants of the human genome are catalogued in the publicly available HapMap databasew1. The SNP in Figure 1, rs3827760, is found in two variants or alleles: at that point in the sequence, one of two bases may be found – either thymine (T) or cytosine (C).
Each circle in the figure represents a single population, and depicts the frequency in that population of the two possible alleles.
The thymine-containing allele is present in all African and most European samples, but is almost completely absent in East Asia and the Americas, where cytosine is most prevalent at that position in the sequence (Sabeti et al., 2007, 2006; Xue et al., 2009).
If we did this comparison for all the other three million SNPs from the HapMap we would see that the distribution of rs3827760 variants among human populations is very unusual. Thus rs3827760 definitely deserves a more detailed look, even though the distribution does not tell us anything about the potential benefit of the variants (their adaptive value), or even whether they are adaptive at all. All we know so far is that for some reason, the original thymine that was present at this position in ancestral human populations in Africa changed to cytosine, and that this change spread through East Asians and Americans. Even the estimates of when this change happened are very imprecise: somewhere between 1000 and 70 000 years ago, all individuals in East Asian populations had the cytosine variant.
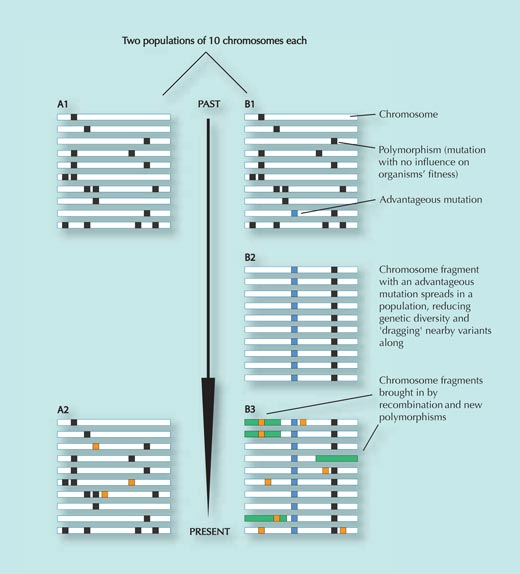
For each individual, one chromosome is shown (e.g. chromosome 9).
Population A does not experience positive selective pressure and remains relatively unchanged over time, apart from
acquiring some random genetic changes that do not influence fitness (orange squares in panel A2; variants which reduce
fitness are removed from the population) – compare panels A1 and A2.
Population B moves to a new environment, however, where it faces new selective pressures. In that new environment,
a particular genetic change (the blue square on panel B1) provides an advantage to the individuals carrying it and
spreads quickly through the population (individuals carrying it leave more offspring). The genetic variants close to
the selected SNP get dragged along with it (the closer two variants are, the smaller the chance that they will be separated
during recombination, when parts of the DNA are exchanged between maternal and paternal chromosomes – see Figure 3).
The result of this rapid spread of a DNA sequence through the population is a reduction in genetic diversity in that region;
most individuals will have the advantageous SNP, together with the genetic variants close to it (compare panels B1 and B2).
This process happens quickly. After some time, however, new genetic changes and recombination events introduce new
variants (green rectangles and orange squares in panel B3). The longer the time since the spread of the original selected
variant, the more difficult it is to detect, because the pattern of reduced diversity (B2) will eventually be masked (B3)
Image courtesy of Jarek Bryk
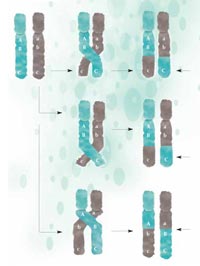
recombination, parts of the
DNA are exchanged between
maternal (green) and
paternal (grey) chromosomes
and passed on to the next
generation in a new
configuration. The closer two
regions are together, the less
likely they are to be
separated during
recombination: A and B are
closer together than B and C,
thus are less likely to be
separated. Click to enlarge
image
Image courtesy of Nicola Graf
How, then, can we decide whether this pattern arose due to positive selection (cytosine conferred an advantage in East Asia and the Americas) or is simply due to chance? To see whether the DNA change (thymine to cytosine) was positively selected, we look at the DNA sequence around the SNP. If the DNA sequence surrounding rs3827760 were similar in all populations, we would have no evidence that the SNP had an effect on the organism’s fitness. If, however, one population (East Asian, for example) were exposed to a selective pressure and rs3827760 contributed to the development of adaptation to that selective pressure, DNA sequences around the SNP would differ between populations. To understand why that is, see Figure 2.
When the DNA sequences around rs3827760 are compared, it becomes obvious that the diversity around the cytosine variants in the East Asian populations is indeed much lower than the diversity around the thymine variants found in the African and European populations (the Americans were not tested). This suggests that positive selection was responsible for the cytosine variant spreading in the East Asian populations. But was this SNP really selected – does it in fact do anything?
Not all changes to the DNA sequence have an effect on protein sequences: most of the SNPs catalogued in the HapMap database either are located in the non-coding parts of the genome (e.g. between genes) or are synonymous – that is, they are located in the coding part of the genome but do not cause a change in the protein sequence encoded (see Figure 4).
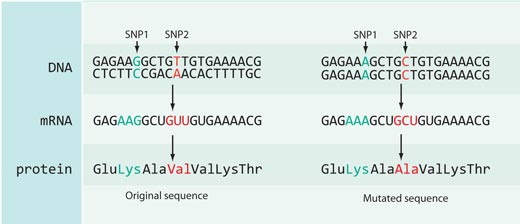
Image courtesy of Jarek Bryk
In the case of rs3827760 we are lucky, because it is located in the coding part of a gene – towards the end of a gene called EDAR, which is involved in the development of hair follicles, sweat glands and teeth. Furthermore, the thymine-to-cytosine DNA sequence change results in a change in the protein sequence: Africans and Europeans (carrying the SNP variant with thymine) have the amino acid valine at position 370 of the protein, whereas East Asians and Americans (with the nucleotide cytosine) have the amino acid alanine. This part of the protein is involved in interactions with other proteins, and mutations there are known to cause ectodermal dysplasias – abnormal development of the teeth, hair and sweat glands – in humans and mice (see Figure 5). This fact strongly suggests that an amino acid change at position 370 may not only change the sequence of the protein but also how the protein behaves, affecting the physical characteristics of the organism itself.
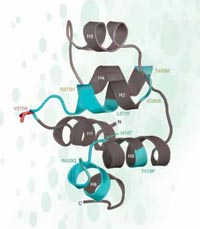
structure of part of the EDAR
protein. Mutations marked in
green and yellow cause
ectodermal dysplasia in
humans. The putatively
selected SNP is marked red
Adapted by with permission
from Macmillan Publishers Ltd:
Nature, Sabeti et al. (2007),
© 2007
To see whether the change in protein sequence really does affect its function, we turn to experiments on the biochemical pathway in which the EDAR protein takes part: a series of reactions which are involved in the development of hair follicles, sweat glands and teeth. When these reactions were carried out in the laboratory, the alanine variant of the protein (found in East Asians and Americans, encoded by the cytosine SNP variant) was found to make the pathway more active than the valine variant (found in Africans and Europeans, encoded by the thymine SNP variant) did. This ties in with comparisons of hair structure, which show that people with the alanine variant have thicker hair than people with the valine variant. For a more direct demonstration, mice were genetically modified to increase the activity of the EDAR pathway. These mice had visibly denser fur with thicker hair, as well as larger salivary glands, than mice with normal EDAR activity (Chunyan et al., 2008; Chang et al., 2009).
Taken together, these findings suggest that the two SNP variants (containing either thymine or cytosine) may affect both the structure and function of the EDAR protein, and may lead to physical differences in humans: differences in hair thickness and, potentially, the size of the salivary glands.
The differences in the DNA sequences that we observe now are historical records of natural experiments, and we can only speculate about the selective pressures that the Asian and American populations were exposed to, which encouraged the spread of the cytosine allele. But the combination of genomic studies, laboratory experiments and animal models makes it possible to test hypotheses about the functional roles of genetic differences between populations or species. Using these approaches, we may uncover the molecular basis of past adaptations in our ancestors and other organisms, highlighting how we adapt to a constantly changing environment.
Glossary
Adaptive value: a trait has an adaptive value if it enables an individual to survive and reproduce better in a given environment than individuals that do not possess this trait. More formally, a trait is regarded as adaptive if it increases fitness.
Fitness: a hard-to-define formal term from evolutionary biology and population genetics; it describes the average number of offspring over one generation that is associated with one genotype compared to another genotype in a population. Thus genotypes that produce more offspring have greater fitness.
Genome: the total DNA of an organism. Usually understood in eukaryotes as the total nuclear DNA, as opposed to including mitochondrial or plastid DNA. For further information, see ‘What is a genome’ on the US National Library of Medicine websitew2.
Positive selection: natural selection is one of the mechanisms of evolution; it describes the different survival and reproduction of individuals in a given environment. Natural selection is called ‘positive’ when it promotes certain traits that help individuals to survive and reproduce better than others.
Selective pressure: a feature of the environment (e.g. temperature; presence of parasites; predation or aggression from members of the same species) that imposes differential survival and reproduction of individuals.
SNP: a single nucleotide polymorphism, or single letter in the DNA sequence that differs between individuals. Pronounced ‘snip’.
References
- Bryk J (2010) Natural selection at the molecular level. Science in School 14: 58-62.
- Chang SH et al. (2009) Enhanced EDAR signalling has pleiotropic effects on craniofacial and cutaneous glands. PLoS ONE 4(10): e7591. doi: 10.1371/journal.pone.0007591
- This article describes the phenotype of various glands of mice with enhanced EDAR signalling, and speculates which traits could have been positively selected in human history. The article is freely available from the journal website: www.plosone.org
- Chunyan M et al. (2008) Enhanced ectodysplasin-A receptor (EDAR) signaling alters multiple fiber characteristics to produce the East Asian hair form. Human Mutation 29(12): 1405-1411. doi: 10.1002/humu.20795
- This article details in vitro studies of EDAR and the transgenic mice, with very nice pictures and photos.
- Pongsophon P, Roadrangka V, Campbell A (2007) Counting Buttons: demonstrating the Hardy-Weinberg principle. Science in School 6: 30-35.
- Sabeti PC et al. (2006) Positive natural selection in the human lineage. Science 312(5780): 1614-20. doi: 10.1126/science.1124309
- This is an excellent overview of various methods used to investigate positive selection from the genomic perspective.
- Sabeti PC et al. (2007) Genome-wide detection and characterization of positive selection in human populations. Nature 449: 913-918. doi: 10.1038/nature06250
- Download the article free of charge here, or subscribe to Nature today: www.nature.com/subscribe
- This paper describes one of the approaches to genome-wide searches for positive selection.
- Xue Y et al (2009) Population differentiation as an indicator of recent positive selection in humans: an empirical evaluation. Genetics 183(3): 1065-77. doi: 10.1534/genetics.109.107722
- This paper contains discussion about EDAR and other similar genes. It is freely available via PubMed Central: www.ncbi.nlm.nih.gov/pmc or using the direct link: http://tinyurl.com/26xte2h
Web References
- w1 – The HapMap project is a partnership of scientists and funding agencies from Canada, China, Japan, Nigeria, the UK and the USA to develop a public resource that will help researchers find genes associated with human disease and the response to pharmaceuticals. See: www.hapmap.org
- w2 – For more information about genomes and the Human Genome Project, see ‘What is a genome’ on the US National Library of Medicine website: http://ghr.nlm.nih.gov/handbook/hgp/genome
Review
Despite all our knowledge of the sequence of the human genome, the precise function of huge segments of it and how and why DNA sequences have changed within populations remain largely undiscovered. Evolutionary adaption in humans has obviously occurred but it is very hard to demonstrate. This article describes how one such change has been identified. Experiments with genetically modified mice have demonstrated how a single base change in the DNA, which changes the amino acid sequence of the protein, leads to an alteration in the structure and function of a protein. This can result in phenotypic variation.
In science lessons, the article could be used when addressing the topics of codon usage and degeneracy; protein structure and function; and population genetics. It could also be used as background reading on variation in the human population or as a starting point for investigating the Sanger Institute and the Human Genome Mapping Project.
The students could discuss evolutionary advantage, with reference to the particular variation described in the article. This could lead on to a discussion of selection, population genetics and Hardy-Weinberg equilibrium. To complement this discussion, there is an excellent activity in Issue 6 of Science in School (Pongsophon et al., 2007).
Suitable comprehension questions include:
- In your own words and giving an example that is not in the text, describe what a SNP is.
- Explain the significance of SNPs.
- Which amino acid does the nucleotide triplet GTT code for?
- Describe the changes that were found in the mice with a genetically modified EDAR pathway, and suggest ways in which the observed changes could be quantified.
Shelley Goodman, UK





