Laying bare our genetic blueprint Understand article
What does the majority of our DNA do? Hundreds of scientists have spent years examining these ‘junk’ sequences, which may hold the key to serious diseases – and much more.

iStockphoto
The Human Genome Project – the sequencing of the human genome – was a major achievement of the past decade: it laid bare the human genetic blueprint, all three billion bases, but the story doesn’t stop there. Deciphering how this sequence is interpreted by our cells is essential to understanding how the genome works. Then, perhaps, we can apply this knowledge to biomedical research and healthcare.

automated DNA sequencing
machine used by the Human
Genome Project to determine
the complete human DNA
sequence. Each peak shows
the presence of a particular
base. The Human Genome
Project identified the 3
billion letters making up our
genome. ENCODE now
provides details of how the
genome works. Click on
image to enlarge.
Image courtesy of Genome
Research Limited
One of the big surprises of the human genome was that only 2% of the genome contains genes, the instructions to make proteins. After accounting for additional bits of the genome such as non-coding RNAs, parts involved in controlling the activity of genes and introns (the sections of a gene’s sequence that are removed before the messenger RNA molecule is translated), a common view was that the rest of the genome had no biological function. As a result, it was often referred to as ‘junk’ DNA.
Going beyond the sequence
Once the human genome was sequenced, it was time to find out whether these sequences really were junk. In 2003, the ENCODE consortium was formed to characterise the non-coding but functional elements of the human genome. The consortium was supported by the National Human Genome Research Institute in the USA and led by the European Bioinformatics Institute (EBI; see box) in the UK. The ENCODE pilot phase ran from 2003 to 2007 and allowed a global network of researchers to test, compare and optimise experimental and computational methods for identifying the active parts in a 1% portion of the genome – essentially sifting through some of the genomic ‘junk’.
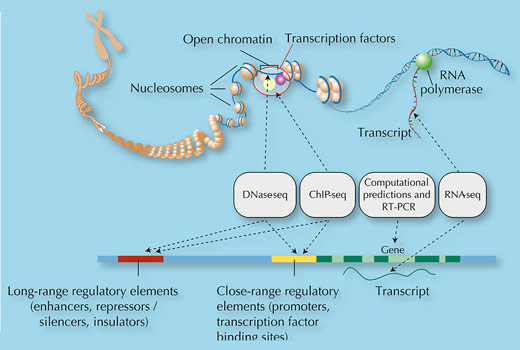
Image courtesy of Ian Dunham
analysed 147 different cell
types to understand
differences in genome
regulation in different tissue
types. This diagram
pinpoints 47 of the 147
different cell types included
in the study. Multiple cell
types were used because
although cells share the
same genome, the way they
use this information differs
between cell types. Click on
image to enlarge.
Image courtesy of Darryl Leja
Their initial results, published in June 2007 (The ENCODE Project Consortium, 2007), gave a tantalising insight into what the genome is doing. For example, the combined data from microarray (see Koutsos et al., 2009) and sequencing experiments showed that the majority of the genome is transcribed, including regions that had been thought to be transcriptionally silent (figure 2). Although the biological roles of most of the transcripts were still unknown, some were shown to be important regulators of gene expression. Overall, this genome snapshot showed that the interplay between genes, regions involved in regulating the activity of genes, and other types of DNA sequences was much more complex than anyone had thought. The data had already started to indicate that the genome contained many forms of active elements and consequently less unused sequence than had been believed.
After successfully testing their approach, the ENCODE researchers then began to examine the entire human genome. This was made easier by advances in DNA sequencing technology and the availability of more precise biochemical assays.
Their analysis systematically mapped features of the genome, just as a map describes a physical landscape and geographical features such as forests, rivers and mountains. In the genome, the ENCODE researchers were looking for features such as regions of the genome flagged with ‘shhhh’ signs (specific types of methyl groups) indicating gene silencing, ‘bind here’ signposts for transcription factors, booster regions to enhance transcription, and DNA modifications that control how the DNA is packaged (figure 3).
Data deluge

project were printed out, the
paper would fill 12 buses.
Image courtesy of
marcus_jb1973 / Flickr
In September 2012, after 5 years of experiments and analysis by 442 researchers from 32 research institutes in the UK, US, Spain, Singapore and Japan, the ENCODE project announced the results of the most detailed analysis of the whole genome to date. The study used about 300 years of computer time to analyse 15 terabytes of data (15 x 1012 bytes), all of which is publicly available. If the data were printed out at a density of 1000 base pairs per cm2, the tower of paper would be 16 m high and more than 30 m long: the equivalent of 12 double-decker buses in volume.
The ENCODE project is an example of what can be achieved by large-scale projects building on the individual contributions of hundreds of researchers, each adding a piece of the jigsaw to produce a complete picture of the genome that could not be achieved by any single organisation.
Bringing the sequence to life
One of the most exciting things that the ENCODE experiments showed is that rather than being predominantly non-functional sequence, our genome is alive with activity: 80% of the genome is actively doing something. Exactly what it is doing remains to be discovered, but certainly 9% of it (and probably much more) is involved in regulating gene expression, controlling when and where proteins are made. The active 80% of the genome contains more than 70 000 promoter regions – the ‘bind here’ sites for transcription factors – and nearly 40 000 enhancer regions – the boosters that control the expression of distant genes.
A massive, 3D control panel

sound engineer, the
expression of genes is under
complex control, with the
human genome containing
over 4 million gene switches.
Image courtesy of Stuart Dallas
Photography / Flickr
Overall, ENCODE identified more than 4 million gene switches dispersed throughout the genome. You could picture the genome as a massive control panel, like a sound engineer’s mixing desk, with lots of switches that turn genes on and off. This information deepens our understanding of gene expression and opens up new opportunities for treating disease. For example, a small change in a gene switch called CARD9 is linked to a 20% increased risk of developing Crohn syndrome, an inflammatory bowel disease. What if you could reset gene switches back to normal, effectively turning off the causes of a disease?
The ENCODE results also shed light on how the genome is organised and the physical interactions occurring within it. The researchers found that these gene switches were in physical contact with the genes they controlled, even though they might be separated linearly by hundreds of kilobases. We tend to imagine the genome as a long, straight line of sequence but in reality it’s all tightly packed in the cell’s nucleus, bringing different parts of the genome in close contact with each other.
Building on the data

a better understanding of
the genetic basis of disease.
Image courtesy of AlexRaths /
iStockphoto
ENCODE provides a detailed map of the genome and opens up whole new areas of science to explore. As Ian Dunham from EBI and lead author on the ENCODE paper explains, “In many cases you may have a good idea of which genes are involved in a disease, but you might not know which switches are involved. Sometimes these switches are very surprising – their location might seem more logically connected to a completely different disease. ENCODE gives us a set of valuable leads to follow to discover key mechanisms at play in health and disease. Those can be exploited to create entirely new medicines, or to repurpose existing treatments.”
As well as knowing which genes are involved in a disease, researchers now know some of the switches involved in regulating how these genes are turned on and off. This will be especially valuable for interpreting the results of population-based studies that identify links between a gene and a disease. By combining ENCODE’s functional analysis of the genome with data from genome-wide association studies, researchers can map the genetic variations that have been linked to disease to the areas of regulatory function, including gene switches, identified by ENCODE. The ENCODE data will enable a better understanding of the genetic basis of disease and support the work of scientists for many years to come.
More about EBI
The European Molecular Biology Laboratory (EMBL)w1 is one of the world’s top research institutions, dedicated to basic research in the life sciences. EMBL is international, innovative and interdisciplinary. Its employees from 60 nations have backgrounds including biology, physics, chemistry and computer science, and collaborate on research that covers the full spectrum of molecular biology.
EBIw2, based near Cambridge, UK, is part of EMBL. It provides data from life science experiments free to the global scientific community, and performs basic research in computational biology. EBI is committed to training researchers in academia and industry to make the most of the incredible amount of data being produced every day in life science experiments.
EMBL is a member of EIROforumw3, the publisher of Science in School.
References
- The ENCODE Project Consortium (2007) Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799-816. doi: 10.1038/nature05874
- The ENCODE Project Consortium (2012) An integrated encyclopedia of elements in the human genome. Nature 489: 57–74. doi: 10.1038/nature11247
- Koutsos A, Manaia A, Willingale-Theune J (2009) Fishing for genes: DNA microarrays in the classroom. Science in School 12: 44-49.
Web References
- w1 – Learn more about EMBL.
- w2 – Learn more about EBI.
- w3 – EIROforum is a collaboration between eight of Europe’s largest inter-governmental scientific research organisations, which combine their resources, facilities and expertise to support European science in reaching its full potential. As part of its education and outreach activities, EIROforum publishes Science in School.
Resources
- Hosted on the Nature website, the ENCODE Explorer enables you to navigate the ENCODE data in its 13 threads. There, you can also download a free poster (20 MB) showing a subset of the ENCODE data.
- In an online video, Nature editor Magdalena Skipper and EBI’s Ewan Birney discuss the ENCODE project.
- The Story of You: ENCODE and the Human Genome presents ENCODE in cartoon format.
- Ewan Birney from EBI, Tim Hubbard of the Wellcome Trust Sanger Institute and Roderic Guigo of CRG present ENCODE in a video (subtitles in Spanish).
- To introduce bioinformatics into your lessons, why not try one of these activities?
- Kozlowski C (2010) Bioinformatics with pen and paper: building a phylogenetic tree. Science in School 17: 28-33.
- Communication and Public Engagement team (2010) Can you spot a cancer mutation? Science in School 16: 39-44.
- To learn more about bioinformatics, see:
- Hayes E (2011) An archaeologist of the genome: Svante Pääbo. Science in School 20: 6-12.
- Pathmanathan S, Hayes E (2007) Nicky Mulder, bioinformatician. Science in School 6: 75-77.
Institutions
Review
This article throws light on one of the latest advances in human genetics: the ENCODE project and how its research was carried out.
When students are introduced to the genetic code, they are often dumbfounded by the fact that only 2% of human DNA actually encodes proteins while the rest is supposedly junk. The ENCODE project has now investigated the function of some of this non-coding DNA, and found that it is not really junk after all.
While discussing the Human Genome Project with students, you can also introduce ENCODE. Providing students with background information on gene regulation, genetic diseases and their treatments, and techniques used in genetics research may be helpful. The article can trigger an interest in bioinformatics among students, and you could encourage them to perform a literature survey on the ENCODE project.
Namrata Garware, India





