An archaeologist of the genome: Svante Pääbo Understand article
Evolutionary geneticist Svante Pääbo tells Eleanor Hayes how he excavates the genome to understand human evolution.
I’d always imagined that archaeologists were to be found up to the knees in mud, digging up bones, pot shards or ancient jewellery. But when I met Svante Pääbo, an ‘archaeologist of the genome’, he wasn’t at all muddy, nor were we out in the open air.

Training Centre at the
European Molecular Biology
Laboratory in Heidelberg,
Germany. Can you see the
similarities to the DNA
double helix? Click on image
to enlarge
Image courtesy of EMBL
Photolab
Instead, we met – appropriately enough – in the shiny new Advanced Training Centre at the European Molecular Biology Laboratory in Heidelberg, Germany. Designed to reflect the DNA molecule, the building has two helical corridors of offices and interconnecting glass bridges to represent the hydrogen bondsw1.
Why the link between DNA and archaeology? “In a way, my colleagues and I do what archaeologists do,” explains Professor Pääbo. “Excavations not in an old cave, but in our genome. We study our DNA sequences for traces of our history, for example to find out where we came from and how we interacted with other forms of humans.”
How, I ask, did he get into this field of research? He explains that an early fascination with Egypt, fuelled by holidays there with his mother, led him to study Egyptology at the University of Uppsala, Sweden. There, however, his romantic dreams were dashed: “We learned about ancient Egyptian word forms, rather than excavating mummies and pyramids in Egypt as I had imagined.” Disappointed, he decided to study medicine instead, and then started a PhD in molecular immunology.
The lure of Egypt persisted, however. In the 1980s, DNA sequence analysis was just beginning. Surely, thought Svante Pääbo, someone must have tried to extract and analyse DNA from Egyptian mummies? “I knew from my Egyptology studies that there are thousands of mummies in museums and hundreds more discovered every year in Egypt.

iStockphoto
But there seemed to be nothing in the literature about DNA extraction from them, so I started working on that myself.” Knowing that his PhD supervisor would not approve, he enrolled the help of his previous Egyptology professor, and then did the laboratory work at night and weekends. The result was every young scientist’s dream: a paper in Nature (Pääbo, 1985).

‘Ötzi’ was found in the Ötztal
Alps
Image courtesy of South Tyrol
Museum of Archaeology
After his PhD and on the strength of his Nature paper, Svante Pääbo moved to the University of California at Berkeley, USA. There he began to realise how difficult it was to avoid contamination when working on ancient human samples – had the DNA he had extracted from the mummies really been ancient Egyptian DNA, or had he been sequencing the DNA of previous anthropologists? He therefore concentrated on extracting DNA from other ancient specimens, including those of the extinct marsupial wolf and of the moa – a giant, flightless bird.
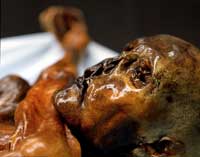
body found preserved in the
ice in the Ötztal Alps, in 1991
Image courtesy of South Tyrol
Museum of Archaeology
When in 1990 he moved to the University of Munich, Germany, to take up a professorship at the age of only 35, he continued working on the DNA of ancient organisms, including mammoths. The advent of the polymerase chain reaction (PCR) enabled scientists to rapidly replicate samples of DNA and thus made it easier to check for DNA contamination by running many identical experiments and using controls. That encouraged Professor Pääbo to start working on ancient human DNA again. This time, he was part of the team looking at the DNA of one of the ancient Egyptians’ European contemporaries: ‘Ötzi’, the 5000-year-old body found preserved in the ice in the Ötztal Alps on the Austrian-Italian border in 1991.
Inspired by the success of the research, which showed that Ötzi’s mitochondrial DNA (see diagram below) was very similar to that of modern central and northern European populations (Handt et al., 1994), Svante Pääbo moved further back in time – about 38 000 years into the past. He wanted to use DNA analysis to investigate human origins.
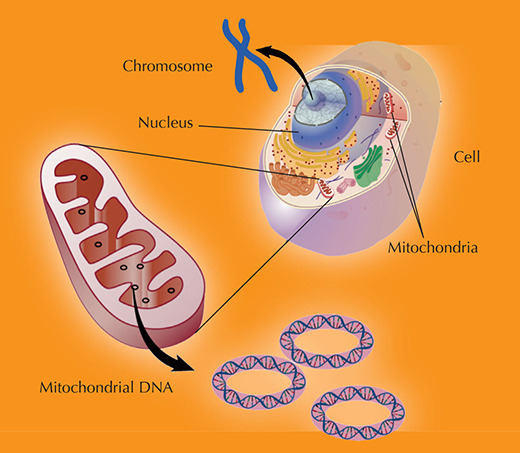
of 23 pairs of chromosomes found in the nucleus (the nuclear genome), as well as a small chromosome found
in the cells’ mitochondria (the mitochondrial genome). These chromosomes, taken together, contain
approximately 3.1 billion (3,1×109) bases of DNA sequence.
Nuclear DNA is inherited from both parents: each parent contributes one chromosome to each pair, so that
offspring get half of their chromosomes from their mother and half from their father.
In contrast, mitochondria, and thus mitochondrial DNA, are passed from mother to offspring
Image courtesy of Darryl Leja, NHGRI / NIH
“One of the big insights from this field in the past 20 years is that modern humans came from Africa rather recently,” Professor Pääbo explains. Although many fewer people live in Africa than outside Africa, “if we look at variations in the DNA sequences of humans, we find that most of the variation exists in Africa and everybody outside Africa is a subset of that variation. It turns out that within the past 100 000 years, a group of Africans left Africa and colonised the rest of the world. So I like to say that from a molecular genomic point of view, we’re all Africans; either we’re living in Africa or we’ve been recently exiled.”

humans encountered
Neanderthals?
Image courtesy of the
Neanderthal Museum, Germany
But modern humans, Homo sapiens, were not the only humans around at the time. Roaming Europe and western Asia (the Near and Middle East) from around 300 000 to 30 000 years ago were our relatives: the Neanderthals, H. neanderthalensis. Ever since the discovery in 1856 of some odd-looking human bones in the Neander Valley near Düsseldorf, Germany, controversy has raged over the fate of the Neanderthals. When modern humans migrated from Africa to Europe, did they kill the Neanderthals? Outcompete them? Or interbreed with them? When Europeans see a picture of a Neanderthal, are we looking at one of our distant ancestors? Svante Pääbo and his colleagues decided to find out, by comparing DNA extracted from the bones of a 38 000-year-old Neanderthal specimen to DNA from various populations of modern humans.
The results were fascinating: non-African modern humans carry some DNA sequences that are similar to Neanderthal sequences but are not found in Africans. In fact, the scientists’ data suggest that between 1 and 4% of the genome of non-Africans is derived from Neanderthals (Noonan et al., 2006).
“The simplest explanation is that when modern humans first left Africa, they came through the Middle East and then went on and colonised the rest of the world. In the Middle East, modern humans interbred with Neanderthals, and their descendants carried the Neanderthal DNA sequences with them – to Australia or Papua New Guinea or the Americas,” explains Professor Pääbo. Only the African populations were unaffected (see diagram below).
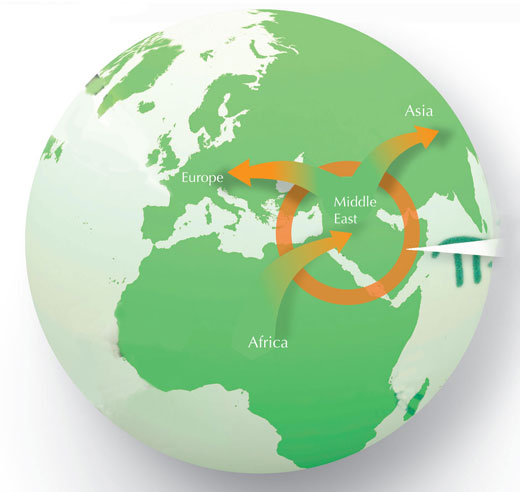
Image courtesy of Nicola Graf

Tyrol Museum of Archaeology /
Kennis / Ochsenknecht
“What’s so fascinating to me is that with DNA sequence analysis, we can answer questions that we cannot address by looking at the skeletons of early humans or the stone tools that they left behind.”
Since 1997, true to his early fascination for human origins, Professor Pääbo has been a director of the Max Planck Institute for Evolutionary Anthropology in Leipzig, Germany.
There, researchers from the sciences and the humanities investigate the history of humankind by comparing the genes, cultures, cognitive abilities, languages and social systems of past and present human populations, as well as those of primates closely related to human beings.
How, I ask him, does he do his research? “My work is in the laboratory, but we collaborate with the people who excavate bones or with the curators of museums that receive the bones. And increasingly, because we can look not just at small parts of DNA but also at entire genomes, it’s crucial to analyse the data with computers.” Svante Pääbo, a hands-on laboratory researcher, finds this frustrating sometimes. “I don’t really know the nuts and bolts of bioinformatics, so I’m in the hands of the people who do. Now, when we train our students, we make sure they learn both sides: young scientists who are primarily working with computers need to spend some time in the laboratory to understand what goes on, and vice versa.”
@EIROforum: ESRF research

One of the most exciting uses of the very intense X-rays produced at the European Synchrotron Radiation Facility (ESRF)w2 is the non-invasive study of fossils. Fossil teeth in particular contain essential information, as they can be used to determine the exact age of a juvenile when it died: by counting daily growth lines in the tooth enamel.
A remarkable finding of recent studies is that Neanderthals grew to adulthood significantly faster than modern humans, H. sapiens, including some of the earliest groups leaving Africa 90-100 000 years ago. The pattern found in Neanderthals, H. neanderthalensis, appears to be intermediate between that found in early members of the genus (e.g. H. erectus) and modern humans (H. sapiens). This suggests that slow development and long childhood are characteristics of H. sapiens, and that the genus Homo shifted from a primitive ‘live fast and reproduce young’ condition to a ‘grow slowly and learn from your parents’ strategy, which has helped modern humans to achieve their current position on our planet.
To find out more, see the press releasesw3 or refer to the original research papers: Macchiarelli et al. (2006) and Smith et al. (2010).
ESRF is one of the members of EIROforumw4, the publisher of Science in School.
Currently, Professor Pääbo is working on an even more distant human relative than the Neanderthals. And at first glance, he and his colleagues had very little to go on: just the tip of a little finger bone, found in southern Siberia. The scientists knew that this bone belonged to some sort of hominid – or human form – but initially they knew very little more about it. What did it look like? How widespread was it? Could it have been a type of hominid found right across Asia while the Neanderthals were limited to more western regions?

Antón / PLoS Biology
The scientists began by sequencing the mitochondrial genome, and found that it was very different from those of both modern humans and Neanderthals. By comparing the DNA sequences of the three groups, they were able to estimate how long ago they started to evolve their separate ways. (To learn how to do this in the classroom, see Kozlowski, 2010.)
Whereas the last common ancestor of Neanderthals and modern humans lived half a million years ago, the mystery hominid diverged from the Neanderthal-human ancestor about a million years ago. That small fragment of bone belonged to a very distant human relative indeed.
This is the first time that a new hominid has been described purely by its DNA sequence but, believes Svante Pääbo, such analyses will become increasingly popular. “This little bone fragment has hardly any information about what the individual looked like, but if it’s well enough preserved, our next step will be to reconstruct the whole genome from it. I think that in future, we will describe newly discovered organisms by their DNA rather than by how they looked.”
This article is based on an interview with Svante Pääbo at the European Molecular Biology Laboratory in June 2010.
References
- Handt O et al. (1994) Molecular genetic analyses of the Tyrolean ice man. Science 264(5166): 1775-1778. doi: 10.1126/science.8209259
- Kozlowski C (2010) Bioinformatics with pen and paper: building a phylogenetic tree. Science in School 17: 28-33. www.scienceinschool.org/2010/issue17/bioinformatics
- Macchiarelli R et al. (2006) How Neanderthal molar teeth grew. Nature 444: 748-751. doi:10.1038/nature05314
- Download the article free of charge here, or subscribe to Nature today: www.nature.com/subscribe
- Noonan JP et al. (2006) Sequencing and analysis of Neanderthal genomic DNA. Science 314(5802): 1113-1118. doi: 10.1126/science.1131412
- Pääbo S (1985) Molecular cloning of ancient Egyptian mummy DNA. Nature 314(6012): 644-645. doi: 10.1038/314644a0
- Download the article free of charge from the Science in School website here, or subscribe to Nature today: www.nature.com/subscribe
- Rau M (2010) Science is a collective human adventure: interview with Pierre Léna. Science in School 14: 10-15. www.scienceinschool.org/2010/issue14/pierrelena
- Smith TM et al. (2010) Dental evidence for ontogenetic differences between modern humans and Neanderthals. Proceedings of the National Academy of Sciences of the United States of America 107(49): 20923-20928. doi: 10.1073/pnas.1010906107
- This open access article is freely available online.
Web References
- w1 – For a virtual tour of the new building at the European Molecular Biology Laboratory (EMBL), Europe’s flagship laboratory for basic research in molecular biology in Heidelberg, Germany, see: www.embl.de/events/atc/tour
- EMBL is a member of EIROforumw4, the publisher of Science in School. To learn more, see www.embl.org
- w2 – An international research centre in Grenoble, France, ESRF produces high-brilliance X-ray beams, which serve thousands of scientists from all over the world every year. To learn more, see: www.esrf.eu
- w3 – The press releases about ESRF research on Neanderthals are available on the ESRF websitew2 or via the direct links http://tinyurl.com/neanderteeth and http://tinyurl.com/neanderkids
- w4 – For more information about EIROforum, see: www.eiroforum.org
Resources
- To find out more about some of Svante Pääbo’s work on primate evolution, see the entertaining fictional dialogue between the ancient Greek philosopher Democritus and his student: www.embl.de/aboutus/science_society/writing_prize/2002_jekely_brochure.pdf
- For more about what our DNA can tell us about human evolution, see the following articles by one of Svante Pääbo’s recent PhD students:
- Bryk J (2010) Natural selection at the molecular level. Science in School 14: 58-62. www.scienceinschool.org/2010/issue14/evolution
- Bryk J (2010) Human evolution: testing the molecular basis. Science in School 17: 11-16. www.scienceinschool.org/2010/issue17/evolution
- For a review of a fictional account of the Neanderthals, see:
- Madden D (2008) Review of Dance of the Tiger. Science in School 8: 68. www.scienceinschool.org/2008/issue8/dance
- To find out how X-ray studies at ESRF can cast a light on the evolution and migration of more distant human ancestors, see:
- Tafforeau P (2007) Synchrotron light illuminates the orang-utan’s obscure origins. Science in School 5: 24-27. www.scienceinschool.org/2007/issue5/orangutan
- For more information about ‘Ötzi’, see the website of the South Tyrol Museum of Archaeology, where the iceman is preserved: www.iceman.it
Review
Cutting-edge science is not just about the future – modern technology can also deepen the understanding of the past. This article about how the study of the genome contributes to archaeological research used in biology lessons to interest students in genetics and evolution. It could also form the starting point of some interdisciplinary research linking biology and history, perhaps using ‘Ötzi’.
Suitable comprehension and extension questions include:
- Using your knowledge about the DNA structure, comment on the architecture of the DNA-shaped building.
- Explain PCR technology.
- Explain the evidence for the claim that “from a molecular genomic point of view, we’re all Africans”.
- Explain how the scientists sequenced the mitochondrial genome. Why did they focus on the mitochondrial rather than the nuclear genome?
The article could also be used as the basis of discussions, for example about:
- The importance of replication of experiments and controls (why PCR was so important)
- The development of scientific knowledge (for an individual or humans generally – see also Rau, 2010)
- The interdisciplinarity of science
- Ethics (despite differences such as skin colour, ‘we’re all Africans’).
Betina da Silva Lopes, Portugal





