Painting life green: GFP Understand article
From jellyfish to arsenic detectors via a Nobel Prize: Sonia Furtado reports on the discovery and development of the green fluorescent protein, and interviews scientists at the European Molecular Biology Laboratory (EMBL) in Heidelberg, Germany, about its applications.
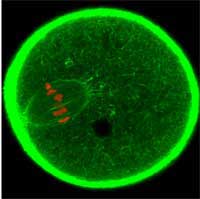
oocyte is dividing. GFP was used
to label the cell’s actin cytoskeleton
green and the chromosomes red
Image courtesy of Jan Ellenberg / EMBL
It started with a jellyfish: transparent Aequorea victoria has spots around its rim that glow green when it is agitated; a behaviour that was first described by scientists in 1955. Studying this ‘oddity’ led to what has been hailed as a scientific revolution, and recently, for three scientists, to a Nobel Prizew1. All because of a single protein, called green fluorescent protein (GFP), which is responsible for the jellyfish’s fluorescence.
In the early 1960s, Japanese scientist Osamu Shimomura discovered aequorin, a jellyfish protein that glows in the presence of calcium. However, aequorin glows blue, while the jellyfish glows green, so something must transform aequorin’s blue light into the jellyfish’s green light. Shimomura discovered this something is another protein: GFP, which absorbs the aequorin’s blue and ultraviolet light and emits green light, giving the jellyfish its glow.
Along came US biologist Martin Chalfie with an idea how to make use of this effect: he reasoned that model organisms could be genetically engineered by attaching the gene for GFP to a specific gene that scientists wanted to study. Then, when the gene of interest was expressed, i.e. when the protein it encodes was produced, it would have GFP attached. This would allow scientists to know when and where a particular gene is turned on: they would just have to shine ultraviolet light onto their organism and look for the green glow.
At the time, two things were apparently missing before Chalfie’s vision could become a reality. The gene for GFP had to be identified, and the mechanism behind GFP’s fluorescence had to be unveiled. Scientists knew that GFP glows because three of its amino acids form a fluorophore, a chemical group that absorbs and emits light. They assumed that, like most naturally fluorescing molecules known at the time, other proteins called enzymes would be needed to fold GFP into the correct shape, and thought that only A. victoria would produce them.
So when biochemist Douglas Prasher identified the GFP gene in 1992, the general consensus was that introducing this gene into other organisms would result in the production of a non-fluorescent version of GFP. However, when Chalfie and his team attached the newly found GFP gene to a bacteria’s DNA, the bacteria glowed green!
It turns out that GFP doesn’t need enzymes to make it glow. Instead, it spontaneously folds into the fluorescent shape, and, biochemist Roger Tsien discovered, the reaction between the amino acids in the fluorophore requires only oxygen, which is readily available in most living cells.
Image courtesy of Typoform / the Royal Swedish Academy of Sciences (RSAS)

the shape of a cylinder, with
the fluorophore at its centre
Image courtesy of Typoform /
the Royal Swedish Academy of
Sciences (RSAS)
Having established exactly how GFP’s fluorophore is formed, Tsien was then able to manipulate this protein. By exchanging different amino acids in different parts of the chain, he developed new versions of GFP which were brighter, absorbed light of different wavelengths, and glowed in different colours: cyan, blue and yellow. And, once a red fluorescent protein was found in coral, Tsien and his colleagues used their knowledge of GFP to make that red fluorescent protein useable as a biological marker too.
Shimomura, Chalfie and Tsien were awarded the Nobel Prize in Chemistry in 2008, “for the discovery and development of the green fluorescent protein”, and scientists all over the world have continued to develop variants of GFP which are now available in virtually all colours of the rainbow.
By now, GFP has become an invaluable tool for scientists all over the world. It does much less damage to cells than chemical fluorescent markers do. After being illuminated for a certain period of time, a fluorophore releases an electron, and after that it will never fluoresce again: it is bleached.
The electrons released in this bleaching process very quickly react with oxygen, forming highly toxic oxygen radicals, which damage cellular components, eventually causing the cells to die. But GFP’s structure acts as a shield, protecting the cell. When the fluorophore releases an electron, the radicals that are formed react within GFP, so they do damage to GFP but not to the cell.
And even though scientists use GFP as a marker by attaching it to a specific protein, different researchers use it to study different processes occurring on completely different scales, tagging anything from groups of cells to individual molecules.
“That’s the beauty of GFP,” says Darren Gilmour, a scientist at EMBLw2: “with it, you can look at all these different scales – you can paint them all with the same paint, you don’t need to change brush.”
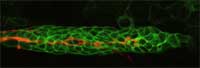
orange, guides other cells
(green) as they migrate along
a developing zebrafish embryo
Image courtesy of Darren
Gilmour / EMBL
In their work on zebrafish embryos, Darren and his group use GFP to tag groups of cells, which they can then follow as the embryo develops, watching how they behave, where they go, and what tissues and organs they ultimately give rise to. The zebrafish embryos Darren studies are transparent, so you’d think it would be easy to see what was happening in them. The problem, Darren says, is that there is too much happening. “It’s an overload. You just can’t focus, you can’t pick out one thing. But with GFP,” he adds, “you can turn out the lights and just focus on a group of cells, or even on a single cell.” (To read more about Darren’s work, see Spinney, 2007).
This ability to follow dynamic processes is also crucial for Francesca Peri, as it allows her to follow the development of her labelled zebrafish embryos under a microscope for days. The alternative would be to sacrifice the animal, slice it and take still pictures. “It would be like trying to understand a football match based on just half a dozen photographs,” says Francesca. “You’d never get the whole picture.” She and her group study microglia, cells which are able to eat dying or damaged neurons. “Using GFP, we can colour-code the different cell types, so microglia will be labelled green, for example, and neurons red. So if we see a red cell inside a green one, we know that a neuron has been eaten to prevent it damaging the rest of the brain tissue.”
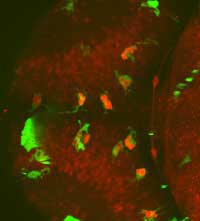
a living zebrafish embryo’s
whole brain, observe interactions
between microglia (green) and
neurons (red) and find neurons
inside microglia (bright red)
Image courtesy of Francesca Peri
/ EMBL
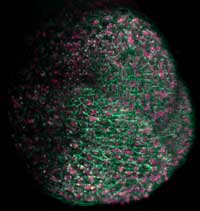
plant’s shoot apical meristem, which
will give rise to the above-ground
part of the plant, fluorescent proteins
were used to label cell membranes
green and nuclei pink
Image courtesy of Ernst Stelzer /
Marcus Heisler / EMBL
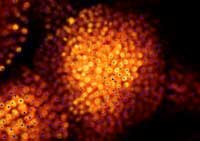
with GFP labelling,scientists can
compare their predictions
(blue) with the real-life location
of cell nuclei (orange)
Image courtesy of Marcus Heisler
/ EMBL
Marcus Heisler and his group use GFP and its variants to study plants. They focus on a plant hormone called auxin, which is transported to the outside of the cell by a carrier that sits on the cell membrane. This carrier can move around the cell, changing the direction in which it sends the hormone. “GFP allows us to follow that very dynamic process in living plants,” Marcus says, “and it gives us good 3D resolution.”
GFP’s reliability is crucial for obtaining such 3D images, says Ernst Stelzer, whose group focuses on developing technologies for 3D imaging over time. “Getting the dye into a thick specimen is always a very serious problem.” he says. Markers injected into the specimens tend not to penetrate very well, so the labelling tends to be uneven: outer layers will be well labelled, but ones near the middle tend to be badly labelled, if at all. “With GFP,” Ernst points out, “we can be really sure that the whole specimen will be labelled, because the dye is produced inside the cells.”
Another EMBL scientist, Rainer Pepperkok, says, “With GFP, we can really do molecular biology in cells, while things are moving, instead of in a test tube.” He and other scientists take advantage of the fact that GFP now comes in many different colours, and exploit a physical phenomenon called fluorescent resonance energy transfer (FRET).
This phenomenon occurs when two fluorescent molecules of different colours – classically red and green – come close to each other. If the green one then receives ultraviolet or blue light, it will absorb that light and transfer some of the light’s energy to the red molecule, which will then emit red light, in a manner similar to GFP glowing green in the jellyfish thanks to the blue light emitted by aequorin. “So if you have a protein tagged with green GFP and another with RFP (red fluorescent protein),” Rainer explains, “if they interact with each other, the red will be brighter and the green will be dimmer.”
Despite all of GFP’s different uses, or perhaps because of them, scientists are still not satisfied. A common request would be GFPs that glow under red or even infra-red light, as this penetrates biological tissues better. “It would also expand the spectrum of available colours,” Jan Ellenberg points out, “which would allow us to tag more proteins and follow them at once.” It may take a new discovery to make this wish come true, as Jan believes the standard way of making GFPs which glow at longer wavelengths – stacking more amino acids onto the fluorophore – is pretty much exhausted.
For his part, Darren says microscopy techniques now need to catch up: “At the moment, we’re at a stage where we could make a zebrafish with five colours, but then we wouldn’t be able to tell them apart with most of the available microscopes.”
Nevertheless, GFP is expanding beyond the realm of science. It is employed in some glow-in-the-dark toys, in glow-in-the-dark fish being sold as pets, and even in bacteria that have been genetically modified to detect arsenic, TNT and heavy metals.
In spite of all these advances, however, the initial mystery remains unsolved: we still don’t know why the jellyfish evolved the ability to glow green in the first place.
References
Spinney L (2007) The great migration. Science in School 7: 20-23.
Web References
- w1 – To learn more about the Nobel Prize in Chemistry in 2008, see: http://nobelprize.org/nobel_prizes/chemistry/laureates/2008
- w2 – Find out more about EMBL and its scientists here: www.embl.org





