Biological crystals: at the interface between physics, chemistry and biology Understand article
Dominique Cornuéjols from the European Synchrotron Radiation Facility introduces us to the world of crystallography. It’s not all shiny diamonds…

/iStockphoto
‘Crystal’ is not a word that immediately comes to mind when thinking about biology. Crystals are better known as magnificent representatives of the mineral world. Gemstones, the shining stars of the underground world, have fascinated us since time immemorial, and the most famous of them, the diamond, has become the symbol of both hardness and eternity.
On the contrary, most biological tissues are soft, and everyone knows that life is not eternal. However, it is possible to isolate the molecules of life, such as proteins, and grow biological crystals from them in the lab. The study of such artificially grown biocrystals has driven – and is still driving – an entire discipline known as macromolecular crystallography.
Crystals and structure
The scientific study of mineral crystals (crystallography) started at the end of the 17th century. At first, it meant describing and measuring the faces and angles of different crystalline structures and classifying them according to their geometric characteristics. Soon, crystallographers proposed that the definite geometry observed at the macroscopic scale be explained by the regular arrangement of very small particles (in fact atoms, molecules or ions), invisible to the naked eye and even under a microscope. With the atomistic theory still in its infancy, this model was extensively debated in the 18th and 19th centuries, without a definitive conclusion.
A series of crucial breakthroughs in physics revolutionised the way we look at matter today:
- In 1895, W.C. Röntgen discovered X-rays. These rays have the extraordinary ability to penetrate objects and bodies, thus revealing their internal features. X-rays found an immediate application in medical imaging, but their nature (as electromagnetic waves) remained an enigma for the next 17 years.
-
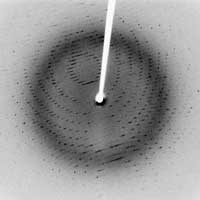
X-ray diffraction pattern of
crystallised 3Clpro SARS protease
Image courtesy of Jeff Dahl; image
source: Wikimedia CommonsIn 1912, Laue discovered X-ray diffraction, when he had the bright idea to put a crystal in an X-ray beam. In a crystal, atoms or molecules are arranged in an orderly, repeating pattern at distinct positions: it is a regular array of atoms. Since atoms act as X-ray scatterers, the regular arrangement of atoms in a crystal produces constructive interference of the scattered rays, which are therefore emitted in specific directions. These emitted rays are recorded on a detector as spots, and the resulting image is called the diffraction pattern. Laue’s experiment proved two things: that crystals consist of a very regular arrangement of atoms, and that X-rays have a short wavelength, in the order of a few tenths of a nanometre – the typical distance between atoms.
- In 1913, the Braggs (father and son) established a relationship – known as Bragg’s law (see Hughes, 2007, for a further explanation of Bragg’s law) – between the wavelength (λ) of the X-rays, the distance (d) between two atomic planes in a crystal, and the angle of incidence (θ) of the X-rays: 2 d sin θ = n λ, where n is an integer. This important discovery marked the beginning of X-ray crystallography, i.e. the possibility of deciphering the structure of a crystal by exposing it to X-rays. The first atomic-resolution structure to be ‘solved’ (in 1914) was that of table salt, followed very closely by that of diamond. Boosted by these early successes, the very powerful technique of X-ray diffraction has provided scientists with the key to the understanding and clarification of the atomic and molecular structures of all sorts of crystals. “It was like discovering an alluvial gold field with nuggets lying around waiting to be picked up,” as the Braggs themselves acknowledged.
At the same time, the first biological crystals were grown, making it possible to study biological molecules using X-rays. The first diffractive image of a protein, obtained as early as 1930, was of an enzyme called pepsin. Soon after that, scientists were able to isolate a virus, crystallise it and show that it did not lose its biological vigour as a result: the tobacco mosaic virus was still infectious for tobacco plants after crystallisation. Macromolecular crystallography was set to go!
Interestingly, not a single biologist was involved in all this early research on complex molecular structures. It was entirely conducted by chemists, physicists and crystallographers, reflecting the fact that during the first half of the 20th century, many scientists from other disciplines took an interest in biology. This is best represented by the book What is Life?, written in 1944 by Erwin Schrödinger, a well-known physicist in the field of quantum mechanics. Molecular biology, which appeared in the 1940s as a merging of biochemistry and genetics, has been – from the very beginning – an interdisciplinary field. Obviously, this nascent discipline has had tremendous help from innovative tools invented by physicists. On a more conceptual level, the idea that life can be explained by simple chemico-physical mechanisms has been very controversial, and many thought that the complexity of the living world could not be reduced to the interactions between biomolecules. Today, structural molecular biology is recognised as a main branch of biology, and is still developing at a very fast pace.
It relies heavily on macromolecular crystallography, taking advantage of the fact that each protein molecule has its own cloud of electrons, which diffracts the X-ray beam used in crystallography. The shape and size of the electron cloud determines the pattern in which the X-rays are diffractedw2 – that molecule’s signal. The many tiny signals obtained from the large number of protein molecules in a crystal add up to a measurable signal.
The resulting diffraction image, taken from several angles of a rotating crystal, is transformed mathematically (this operation is called a Fourier transform) into an electron density map of the protein, which represents the electron cloud of the protein. With the help of computer modelling and refinement techniques, the sequence of amino acids of the protein can be fitted into this electron cloud to determine the three-dimensional arrangement of atoms in the protein, which is the final structure.
But why is their three-dimensional structure so important for the study of proteins and other biological molecules?
Structure and function
Our hands and eyes, like other anatomical features of the plant and animal kingdoms, have been shaped through evolution to meet the needs of life. In a similar way, the microscopic structure of each subcellular organelle and biological macromolecule is intimately linked to its function. Molecules with the right shapes are responsible for turning genes on and off, catalysing the complex chemistry of life, defending against cellular invaders, and flipping the switches that initiate cell division and control development.
Image courtesy of geopaul / iStockphoto
The importance of molecular structure for an understanding of function is best exemplified, of course, by DNA. The simple and beautiful double-helical, base-paired structure of DNA immediately made genetics intelligible in chemical terms. Genes, the previously mysterious factors that controlled the inheritance of particular traits, were segments of the DNA molecules that could be spooled out of solution at the end of a rotating glass rod, like cotton candy on a stick (see Madden, 2006, for a simple DNA purification classroom protocol), thus producing a fibre that could be studied by X-ray diffraction. The determination of the remarkable but simple structure of DNA marked a milestone in structural biology.
By contrast, the study of the structures of proteins has not yielded a simple and all-encompassing explanation of protein structure and hence function. To this day, and despite knowing the structures of about 45 000 different proteins, we are still unable to establish a set of general rules that would allow us to predict a protein’s three-dimensional structure from the amino-acid sequence of its polypeptide chain. Proteins fulfil a much wider range of biological functions than DNA does, and functional diversity has dictated structural diversity.
A bottleneck in structural genomics: protein crystallisation
In comparison with molecular genetics, progress in research on protein structure has been painfully slow, partly because of the simple technical problem of obtaining protein crystals which are large enough to use for crystallographic analyses and which diffract X-rays well enough to allow the structure to be determined with a high (atomic) resolution. Furthermore, although they look like crystals of small molecules, such as cooking salt crystals, protein crystals are much smaller and generally very fragile.
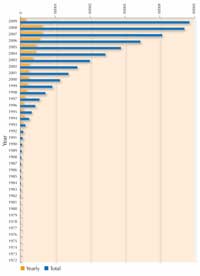
structures solved by X-rays.
Note: searchable structures
vary over time as some become
obsolete and removed from the
database. Click to enlarge image
Protein crystallisation has therefore always been a hit-and-miss business with no predictive theory. Some proteins crystallise readily, others stubbornly refuse to produce suitable crystals; some investigators seem to have ‘green fingers’, like good gardeners, and can grow crystals where others fail. As a result, protein crystallisation has sometimes been felt to be more of an art than a science.
For each new protein, scientists must screen a large number of conditions to find the particular circumstances under which it will crystallise. Variables that can be changed in the conditions are, for example, protein concentration, temperature, pH, and the concentration of a wide range of precipitation agents that may be used in combination with various salts. To try out the best conditions for crystallising a protein in a classroom experiment, and have your results analysed by X-ray diffraction in a real crystallography lab, see Blattmann & Sticher (2009) in this issue. Because protein crystallisation has posed so many difficulties, until recently the most studied proteins have been those that crystallise easily, and which can be produced in large quantities, rather than the most interesting ones. However, much progress has been made in the last decade as can be seen in the figure showing the growth in protein structures solved by X-ray crystallography since 1983. This spectacular progress is due to improved techniques in three areas: crystal preparation, synchrotron X-ray crystallography and software development.
Journey of a protein from lab to lab
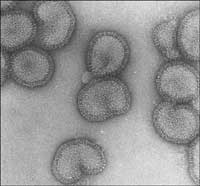
electron microscope
Image courtesy of Rob Ruigrok/
UVHCI
To illustrate how a protein’s structure is solved using today’s state-of-the-art instruments, we will look at how scientists identified the structure of one of the influenza virus’ proteins, the polymerase. A group of scientists from the European Molecular Biology Laboratory (EMBL Grenoble outstationw3, France) and from the Unit of Virus Host-Cell Interactionw4 (UVHCI, Grenoble) have been studying this protein, which is involved in the mechanism that the virus uses to take over key processes in the human cells it infects (see Ainsworth, 2009, in this issue, for details of the study and its findings). For this project, the scientists made use of the Partnership for Structural Biologyw5 (PSB), a collaboration to decipher structures of biological molecules with high medical interest.
Cloning and expression (at the UVHCI, EMBL and PSB)
Once the protein had been selected for study, its corresponding gene was amplified, i.e. cloned into a special expression system. This allows large quantities of protein to be produced using a host system, usually bacteria.
Purification (at the PSB) and quality assessment (at the IBS)
The bacterial cells were then ‘harvested’ by centrifugation, and cell debris and possible contaminants such as nucleic acids were removed. The protein was then subjected to a lengthy but crucial multistep purification process, since at least 95% purity is desirable for crystallisation. Protein quality was assessed at the Institut de Biologie Structurale Jean-Pierre Ebelw6 (IBS) by mass spectrometry (for a short introduction to mass spectrometry, see Wilson & Haslam, 2009, in this issue), and a sequencer was used to check that the purified protein was the one that was wanted: the influenza virus’ polymerase.
Crystallisation (at the PSB)
Scientists started attempting to crystallise the protein by using multifactorial screens. In other words, they exposed different concentrations of the protein to different crystallisation agents, buffers, temperatures, and so forth. Known as the microbatch method (see image below), this is designed to obtain maximum information on the protein one wishes to crystallise while using a minimal amount of sample.
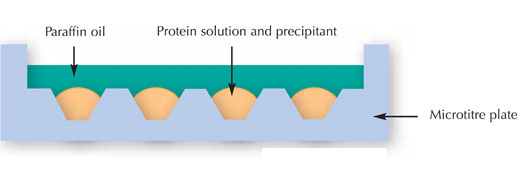
Image courtesy of Nicola Graf
X-ray diffraction on a synchrotron beamline and data collection (at the ESRF)

Image courtesy of ESRF

place on an ESRF beamline
Image courtesy of ESRF
Once they had obtained crystals of the polymerase, the scientists cryogenically preserved them in liquid nitrogen and transported them to a beamline at the European Synchrotron Radiation Facility (ESRF)w1. There, the crystal was fixed on a goniometer head – this is usually done by a robot – and exposed to synchrotron X-rays, which are extremely intense. A goniometer is an instrument that allows an object, such as a crystal, to be rotated to a precise angular position.
The goniometer head is rotated in the X-ray beam, in order to produce a maximum number of reflections, or diffracted beams. This produced enormous quantities of data, as is typical of data collection at synchrotron sources, so the actual structure of the protein was then determined automatically, using a set of software packages specifically developed for the purpose.
Model building, map fitting, refinement and validation (at the UVHCI and EMBL)
Based on the collected and processed data, an atomic model of the polymerase was built and compared with the electron density map. The model was then iteratively refined to best fit the observed data, thanks to powerful software and interactive molecular graphics. After model validation, the structure was finally published and deposited in the publicly accessible Protein Data Bankw7.
The ESRF: one of the most intense X-ray sources in the world
The ESRF is a good example of a large facility operating day and night for the benefit of thousands of users from all over the world. A ‘user’ is a scientist, usually part of a larger team, who occasionally needs a powerful tool to obtain information on a sample of interest (a piece of material, a protein crystal, a fossil, or a catalytic reaction, for instance). Most users travel to Grenoble a couple of times a year to collect data at the ESRF.

Facility
Image courtesy of P. Ginter/ESRF
As a third-generation source, ESRF produces extremely intense X-rays, called synchrotron radiation. These X-ray beams are emitted by high-energy electrons (6 GeV) which circulate in a large ‘storage ring’ measuring 844 metres in circumference. The synchrotron X-rays are very collimated, somewhat like laser beams (the rays of collimated light are nearly parallel).
The X-ray beams are directed towards the beamlines, which surround the storage ring in the experimental hall. Each of the 42 beamlines at the ESRF is specialised in a specific technique or type of research. For around 10 of them, this speciality is protein crystals. The beamlines at the ESRF are becoming ever more automated, making them easier to use and, recently, granting scientists remote access. This allows users to drive their synchrotron experiments without physically leaving their home laboratory. The crystals are shipped rather than personally taken to the ESRF, even if the scientists go there themselves, because current security restrictions make it difficult to travel with sensitive biological samples.
Synchrotron radiation accounts for about 80% of the macromolecular crystal structures currently deposited in the Protein Data Bankw7 (in 1995, only 17% of these came from synchrotron data, see image above). ESRF produces some 20% of this total.
References
- Ainsworth C (2009) Outmanoeuvering influenza’s tricks. Science in School 11: 25-29.
- Blattmann B, Sticher P (2009) Growing crystals from protein. Science in School 11: 30-36.
- Hughes D (2007) Taking the stress out of engineering. Science in School 5: 61-65.
- Madden D (2006) Discovering DNA. Science in School 1: 34-36.
- Schrödinger E (1944) What is Life? Cambridge, UK: Cambridge University Press
- Wilson A, Haslam S (2009) Sugary insights into worm parasite infections. Science in School 11: 20-24.
Web References
- w1 – For more information on the ESRF, see: www.esrf.eu
- w2 – More information on the theory of protein crystallisation and X-ray diffraction can be found here: www-structmed.cimr.cam.ac.uk/Course
- w3 – To find out more about the EMBL outstation in Grenoble, see: www.embl.fr
- w4 – To learn more about the Unit for Virus Host Cell Interaction, see: www.uvhci.fr
- w5 – The Partnership for Structural Biology (PSB) is a collaboration between EMBL, the Institut de Biologie Structurale, ESRF and the Institut Laue-Langevin – the world’s leading neutron source. The PSB provides an integrated structural biology environment with a portfolio of state-of-the-art techniques on one site: cloning and expression, crystal production, physico-chemical-biochemical characterisation, X-ray and neutron crystallography, nuclear magnetic resonance (NMR), electron microscopy and tomography, small angle X-ray and neutron scattering, mass spectroscopy and advanced light microscopy. See: www.psb-grenoble.eu
- w6 – To find out more about the IBS, see: www.ibs.fr
- w7 – For the website of the Protein Data Bank, see: www.rcsb.org/pdb
Resources
- Abad-Zapatero C (2002) Crystals and Life: A Personal Journey. La Jolla, CA, USA: International University Line. ISBN: 9780972077408
- Blow D (2002) Outline of Crystallography for Biologists. Oxford, UK: Oxford University Press
- Branden C, Tooze J (1991) Introduction to Protein Structure. New York, NY, USA: Garland
- Michette A, Pfauntsch S (1996) X-rays: the first hundred years. Chichester, UK: John Wiley & Sons
- Wood EA (1972) Crystals – A Handbook for School Teachers. Chester, UK: International Union of Crystallography
- For a website with crystal growing experiments, see: www.sciencecompany.com/sci-exper
- For the website of the International Union of Crystallography, see: www.iucr.org





