Systems biology in the classroom? Understand article
Systems biology is one of the fastest growing fields in the life sciences. But what is it all about? And does it have a place in the classroom? Les Grivell from the European Molecular Biology Organization (EMBO) in Heidelberg, Germany, investigates.
If you type the phrase ‘systems biology’ into the search box of the main biomedical literature database PubMedw1 and restrict the output to any year before 2000, your query will return only a handful of hits. Do the same for 2008, and the outcome will be links to several hundred publications dealing with this rapidly growing area of biological research. Do the same kind of search in Google and compare the outcome (around 36.9 million hits) with those for areas like high-energy physics (about 11.4 millions hits), or aerospace engineering (around 2.5 million hits). Assuming that in each case the number of hits is a reflection of the ongoing activity in the respective area, systems biology is really quite an active new kid on the block.
But what is it?

tube radio
Image courtesy of Ermin
Gutenberger/iStockphoto
Cynics might reply that ‘systems biology’ is just another buzzword – one more way for researchers to tap into new sources of funding. Physiologists might say that it is nothing new; just a high-tech way of doing what they have always done or have attempted to do. Systems biologists themselves will tell you that it is a radically new way of thinking about biology. Instead of exploring the characteristics of isolated parts of a cell or organism, as biochemists and molecular biologists have done for many years, systems biologists focus on the whole system.
Let’s look at a radio as an analogy: a radio transforms electric waves into sound waves, but we don’t know exactly how. While molecular biology took the radio apart to identify its parts, systems biology will now try to understand how the parts work together to achieve a function.
Consider, for example, the ability of bacteria to detect and swim in the direction of a source of nutrients (chemotaxis). The molecular biologist will try to characterise the individual components of the chemotaxis machinery and how each of them works – by purifying them and the genes that encode them, and by studying the effects of mutations on each component. In contrast, the systems biologist wants to understand how the highly complex swimming and tumbling pattern of the cellsw2 is controlled – by looking at the impact and interactions of as many components in the system as possible. The systems biologists, therefore, may study everything from the first contact between nutrients and the relevant receptors in the bacterium, through the entire signal transduction pathway, to the mechanism that controls the rotation of the bacterial propulsion motor – its flagellum.
long, thin structures called
flagella to propel themselves.
These flagella form bundles
that rotate counter-clockwise,
creating a torque that causes
the bacterium to rotate
clockwise
Image courtesy of Nicolle Rager
Fuller, National Science Foundation;
image source: Wikimedia Commons
According to Leroy Hood, the founder of the world’s first dedicated Institute for Systems Biologyw3 in Seattle, USA, systems biology can be defined through six essential features:
- Global measurements: scientists measure dynamic changes in all genes, mRNAs and proteins, rather than in individual genes or mRNAS or proteins.
- An integration of data types: information about DNA, RNA, proteins and their interactions are computationally and mathematically integrated.
- Dynamic measurements rather than static ones: across different developmental, physiological, disease and environmental areas.
- The research is discovery-driven and hypothesis-driven, rather than only one or the other.
- The measurements made are quantitative, rather than just qualitative (you want to know how much more of a protein is produced under certain circumstances, rather than just that there is more).
- An interactive cycle of data: data → model → prediction → verification → modification → data.
At the molecular level, systems biology often makes use of high-throughput technologies, such as massive DNA sequencing, and cell- or tissue-wide RNA, protein and metabolite analysis, to assemble comprehensive data sets that characterise the system being studied. The ways in which gene expression or metabolites change with time, or in response to genetic mutations and/or stimuli from the environment, are then used to construct computational models that are able to predict behaviour and thereby to better understand the molecular principles and strategies that underlie such changes.
This emphasis on a system as a whole is a significant one, since it marks the reversal of a strongly reductionist approach to research that started with the earliest biochemical studies on isolated enzymes at the end of the 19th and the first half of the 20th centuries (Cornish-Bowden & Cárdenas, 2005). The reductionist approach was necessary in this period: little progress could have been made without isolating and studying the properties and behaviour of components of cells individually. However, it is becoming increasingly clear that the behaviour of a single cell, or of populations of cells, is the result of a complex mix of interactions that feeds both upwards to higher levels of organisation and back downwards to individual molecules or their complexes in such cells.
Kaulitzki/iStockphoto
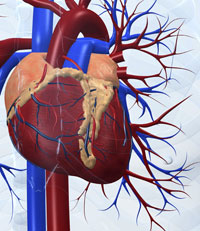
A good example of this kind of complexity is the systems biology studies that have led to the first models of a human organ – the virtual heartw4 (Noble, 2007). Here, biophysical and biochemical studies on specific ion channels in heart cells led first to models of the behaviour of single cells, then to linked models of two- and three-dimensional blocks of tissue in the heart’s atrium and ventricle, and eventually to the simulation of the electrical and mechanical behaviour of the beating heart as an entire organ, in which every cell plays its specific role in concert with its neighbours.
The results of the PubMed searches mentioned earlier may give the impression that systems biology suddenly appeared sometime between 1999 and 2000 and developed rapidly. In reality, systems biology has existed in one form or another for much longer and under different labels. The physiologists cited earlier on could, with good justification, be regarded as the forerunners of the field, since physiology is defined as ‘the study of living organisms and their parts’ and thus is, like systems biology itself, inherently integrative (Strange, 2005).
So what can systems biology tell us? Ultimately, of course, this kind of research offers an understanding of the system being studied, whether that is a relatively simple network of interacting molecules, a cell, a tissue, or an organ.
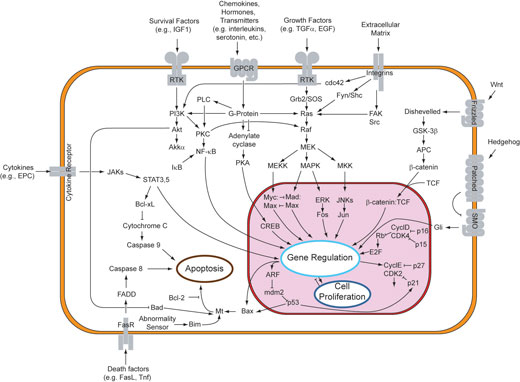
At the level of the individual cell, Cheong and Levchenko (2008) analyse the recent data compiled on the NF-kappaB pathway. This molecular signalling cascade is widely used by cells in the inflammatory response to infection. A wealth of data had been collected on the individual components of the system over the past few decades, but now systems biologists are integrating them into quantitative computer models of the entire pathway within a cell, and then testing experimentally if the predictions made by the model are correct. The results have led to remarkable insight into the underlying, highly complex molecular circuitry that cells use to detect and combat infection. This knowledge will eventually help us understand why individuals differ in their inflammatory responses and thereby should lead to more effective treatment.
At the level of a whole human, Nicholson (2006) proposes a systems approach to metabolism including gut flora. Cellular metabolism is very much a game of chance in which metabolites or drug molecules interact with enzymes and other molecules in a chance fashion. These interactions can result in many outcomes, some of which may cause cellular damage. And this is only for one cell – imagine how complex it is to predict the metabolic state of an entire human being! It is only by study and mathematical modelling of the system as a whole that we can hope to understand the complexity of such responses and develop therapies that are exactly tailored to the system state of any particular individual.
Should systems biology be included in the school curriculum? In my opinion – yes. By this I do not imply that students need to have access to high-throughput microarray or proteomics facilities. Rather, I think it important that they are exposed to some of the basic principles of systems biology, and that above all, they are taught to realise the limitations of the reductionist approaches that have dominated biological research for so long.
Public domain image; image source: Wikimedia Commons
Starting with questions like “What is a gene?”, “How many genes do you need to make a minimal self-maintaining organism?” and “How can I make a biological clock?”, it is possible to introduce typical systems concepts. These include, for example, non-linearity of biological systems, a broad, but important concept: many metabolic and signalling pathways are organised in a cyclic, non-linear fashion. There are negative and positive feedback loops within a cell, and more often than not they will interact with each other at several levels. Besides, the relationships between an input into a biological process and its outcome are often non-linear. So it is quite difficult to predict off the top of your head what will happen if you tweak one component of a pathway to be slightly more or slightly less active, and how this will influence all the other components.
Modularity is another important concept: that is, biological systems are complex, but they can be regarded as networks of smaller and simpler units (modules) that perform defined functions. Other central themes of a system are robustness (continued function despite genetic or environmental perturbations) and evolvability (the potential for change).
The international Genetically Engineered Machine competition (iGEM)w5 challenges university students to put a number of these systems biology principles into practice through the design and use of standardised, biological components. The register of these componentsw6 is a fascinating web resource that also shows that systems and synthetic biology is fun! Alongside an intriguing BacteriO’Clockw7 – a simple test tube containing modified bacteria that change colour according to the time of day (Paris team) – current iGEM team projects include the engineering of Lactobacillus to produce yoghurt that cleans your teethw8 (MIT team), a bacterial biosensor that can be directly integrated into an electrical circuitw9 (Harvard team) and an E. coli cell that glows when it detects pathogenic bacteria in drinking water (Sheffield team).
Finally, upcoming generations need to be made aware that there are tremendous opportunities for tackling a wide range of intriguing problems about the living world that will be of utmost importance to society. Systems biology requires systems biologists and there is a real need for scientists in the disciplines of physics, computer science and biology to work together to develop the field to a stage at which it can begin to return benefits to society as a whole.
Acknowledgement
I am very much indebted to Dr Thomas Lemberger (EMBO), both for his comments on this article and for a variety of other discussions about systems biology.
References
- Cheong R, Levchenko A (2008) Wires in the soup: quantitative models of cell signalling. Trends in Cell Biology 18: 112-118.
- Cornish-Bowden A, Cárdenas ML (2005) Systems biology may work when we learn to understand the parts in the terms of the whole. Biochemical Society Transactions 33: 516-519.
- Nicholson JK (2006) Global systems biology, personalized medicine and molecular epidemiology. Molecular Systems Biology 2: 52.
- Noble (2007) From the Hodgkin-Huxley axon to the virtual heart. Journal of Physiology 580(1):15-22.
- Strange K (2005) The end of “naïve reductionism”: rise of systems biology or renaissance of physiology? American Journal of Physiology 288: 968-974.
Web References
- w1 – To access PubMed, visit: www.ncbi.nlm.nih.gov/pubmed
- w2 – You can find movies on bacterial motility here: www.microbiologybytes.com/video/motility.html
- w3 – For more information on the Institute for Systems Biology, see: www.systemsbiology.org
- w4 – To watch a video of Denis Noble illustrating the principle of systems biology using the virtual heart, see: http://videolectures.net/eccs07_noble_psb
- Find out more about the virtual heart in Noble D (2008) The Music of Life: Biology Beyond Genes. Oxford, UK: Oxford University Press. ISBN: 9780199228362
- Take a look at the virtual heart website from Cornell University (not Denis Noble’s version, which is not online, but a very good one nevertheless): http://thevirtualheart.org
- w5 – To learn more about the iGEM competition, see: http://2008.igem.org/Main_Page
- w6 – The website of the BioBricks register of components to be used in the iGEM competition can be found here; http://partsregistry.org/Main_Page
- w7 – For an explanation and video of the BacteriO’Clock, see: http://2008.igem.org/Team:Paris
- w8 – For more information about the iGEM MIT team’s ‘biogurt’ that cleans your teeth, see: http://2008.igem.org/Team:MIT
- w9 – To find out more about the Harvard team’s ‘bactricity’ project for iGEM, see: http://2008.igem.org/Team:Harvard
Resources
- In an insightful historical overview of the evolution of systems biology, Westerhoff and Palsson (2004) show how ideas on molecular and cellular self-organisation were subsequently extended to modelling and quantitative analysis of metabolic networks. These small-scale approaches constitute important preludes to the development of present-day systems biology.
- Westerhoff HV, Palsson BO (2004) The evolution of molecular biology into systems biology. Nature Biotechnology 22: 1249-1252.
- See the blog ‘What is systems biology?’ at http://blog-msb.embo.org/blog/2007/07/what_is_systems_biology_3.html
- For a review of a more reductionist but nonetheless cutting-edge approach to biology – protein crystallography – see:
- Cornuéjols D (2009) Biological crystals: at the interface between physics, chemistry and biology. Science in School 11: 70-76.
Review
Physiology considers how biological systems work. This article describes how the molecular approach to biology demonstrates how cells and even systems work together to perform systems functions. The virtual heart model on the cited website is worth watching, as it illustrates how the cells perform as a system. The iGEM competition website can be accessed to investigate novel applications of genetic engineering and systems biology. This could provoke some interesting discussion and may motivate some students to come up with their own ideas.
The article could be used to discuss the following topics:
- Heart structure and function
- Genetic engineering: social, ethical, and commercial applications (of products such as tooth cleaning yogurt!)
- Physiology: integration of systems.
Possible comprehension questions to ask the students include:
- What is the purpose of systems biology?
- State what is meant by a molecular signalling cascade.
- Do individuals exhibit the same inflammatory responses?
- Explain how systems biology could be useful in predicting an individual’s response to drug therapies.
Shelley Goodman, UK





