Of Roman roads, train yards and inspectors: recent discoveries in RNA research Understand article
RNA is a crucial biological molecule that is seldom mentioned in detail in textbooks. In the first article in a series, Russ Hodge describes some exciting recent research on RNA.

Florian Raible looking for the
fastest and slowest evolvers
Image courtesy of EMBL
Photolab
Imagine a Roman in ancient times, standing on a summit, surveying the landscape as he plots the course of a new road. Two thousand years later his roads are still visible as they cross European cities and the countryside, as straight as if drawn by a ruler, deviating only in the face of the most stubborn obstacles. In the 1950s and 1960s, scientists mapped out a similar sort of route for biology as they described the connections between different types of molecules in the cell.
Imagine a Roman in ancient times, standing on a summit, surveying the landscape as he plots the course of a new road. Two thousand years later his roads are still visible as they cross European cities and the countryside, as straight as if drawn by a ruler, deviating only in the face of the most stubborn obstacles. In the 1950s and 1960s, scientists mapped out a similar sort of route for biology as they described the connections between different types of molecules in the cell.
Their plan is known as the central dogma of molecular biology: “DNA makes RNA makes proteins,” as James Watson scribbled in one of his notebooks when he and Francis Crick began their assault on the structure of DNA. With the dogma, the two men proposed a new relationship between the cell’s major kinds of molecule – a one-way flow of information from nucleic acids to proteins. Soon after their discovery of the double helix, Crick gave lectures in which he presented the dogma as a grand research plan for molecular biology (Crick, 1970): scientists should devote themselves to uncovering the exact cellular mechanisms underlying the transformations.
By the 1970s, the basics of how RNAs and proteins were created were understood, but it was becoming obvious that a single straight road could not describe the chemistry of the cell. Just as a score of modern side-streets branch off those ancient Roman roads, many deviations have been found on the route between genes and proteins. At first these seemed to be rare exceptions to the dogma; in the meantime, they have been recognised as common and crucial in most biological processes in complex organisms. Some of the findings are so new that they have not yet made it into textbooks and may be unfamiliar to teachers. This article is the first in a series for Science in School that will discuss some of this recent research.
Some of the most exciting findings involve RNA, once thought to be little more than a means to an end in making proteins. Over the last decade these molecules have come to be seen as crucial players in the cell’s control of the information in its genome. Understanding how RNAs are handled has led to insights into disease and powerful new kinds of biotechnology, including efforts to use RNAs in therapies to counteract the effects of defective genes.
Alternative splicing
The RNA of the central dogma is messenger RNA, which serves as a template for making proteins. In eukaryotes, the RNAs usually have to be processed before achieving the messenger form. Most freshly made RNAs are like long trains with many boxcars (freight wagons), whose cargo needs to be sorted to fit the needs of different customers. Empty cars are detached, and the cars in front and behind are rejoined. The cellular parallels of the empty cars are called introns, large regions within genes that do not encode proteins. Protein-encoding DNA sequences are called exons. Often one exon contains the recipe for one module within a protein. Even these are not always used. A customer might not want all the wares on a train, so some of the boxcars that contain cargoes may be removed along with the empty ones. And a cell might not need all of the modules of a particular protein, so various exons might be combined to produce different forms.

Image courtesy of EMBL Photolab
This process, called alternative splicing, was discovered independently in 1977 by Richard Roberts of New England Biolabs, USA, and Phillip Sharp of the Massachusetts Institute of Technology, USA (Berget et al., 2000). The finding was so significant that it earned them the Nobel Prize in Physiology or Medicine in 1993w1. At the time splicing was thought to be rare; Sharp estimated that only about 5% of human genes were likely to undergo alternative splicing. The complete human genome sequence has revealed that the average gene contains 8.4 introns, all of which have to be removed through splicing. Although some human RNAs are probably always spliced the same way, scientists now estimate that at least 75% undergo alternative splicing.
Human genes have considerably more introns than those of most other organisms; the average for flies and other insects lies between 2.4 and 5.4 introns per gene. (Even so, the current world-record holder for alternative splicing is a fruit fly gene called dscam, which can generate 38,016 distinct proteins.) Some researchers believed that the high number of human introns meant that genes were becoming more complex over time. However, a recent study by the groups of Detlev Arendt and Peer Bork at the European Molecular Biology Laboratory (EMBL) in Heidelberg, Germany, shows that the ancient common ancestor of insects and vertebrates almost certainly had genes more like those of humans, with more introns (Raible et al., 2005). Genes have become more streamlined over the course of evolution in flies and other quickly reproducing species.
There are also interesting differences between species when it comes to the total length of introns compared to exons within genes. In the genes of the worm Caenorhabditis elegans and many other species, introns and exons contain about the same number of ‘letters’ (nucleotides). The situation in humans is very different: introns in a single gene frequently total tens of thousands of nucleotides and are, on average, five times the length of the exons. This may have an impact on the evolution of human genes, as revealed in a study by Cristian Castillo-Davis of Harvard University, USA, and Eugene Koonin and Fyodor Kondrashov of the National Center for Biotechnology Information, USA (Castillo-Davis et al., 2002). Transcribing RNA is a slow and energy-expensive process: making RNAs from a single gene with huge introns can require several minutes and thousands of molecules of ATP. The authors found that the introns of frequently used genes are on average 14 times shorter than those of rarely used genes. Their conclusion: natural selection has been shortening introns in the most common genes, saving time and energy.
Once the cellular machinery needed to carry out alternative splicing evolved, it could be put to use in many different ways. Mixing and matching modules produces proteins that behave differently. They help to create diverse types of cells and figure prominently in the development of different tissues. Alternative splicing of an RNA called Slo in the ear of the chicken improves the bird’s hearing by giving it cells sensitive to different frequencies of sound. In flies, three critical proteins are spliced differently in males and females, creating important differences between the sexes. Although females have two X chromosomes and males only one, females don’t produce twice the amount of proteins from the genes on the chromosome, thanks to the differences in these proteins. Diane Lipscombe and her colleagues at Brown University (Rhode Island, USA) have found that alternative splicing is especially common in the brain of mice and other mammals (Lipscombe, 2005). Some of the molecules are crucial to memory and learning.
Splicing is a factor in a wide range of diseases. Half of the people who suffer from neurofibromatosis, a severe genetic disease in which tumours develop alongside nerves and other tissues, have mutations that change the splicing of RNAs made from the neurofibromin gene. Patients with beta-thalassaemia suffer from anaemia as a result of non-functional beta-globin proteins in their red blood cells; this severe disease is caused by mis-splicing of the responsible gene. Other examples are changes to the BRCA1 gene (linked to breast cancer) and CFTR gene (leading to cystic fibrosis). It is estimated that about 50% of disease-related mutations in exons affect the way RNAs are spliced. Tumours and neurodegenerative diseases are often accompanied by unusually spliced RNAs that are not normally found in healthy tissues.
Quality control
In 1979, Regine Losson and François Lacroute of the CNRS in Strasbourg, France, discovered that the cell has a system to inspect RNAs and carry out quality control (Losson & Lacroute, 1979). Nearly three decades of research have shown that the system is not perfect, but it manages to protect eukaryotic cells from the dangerous effects of most mutations. Defects in genes that change the shape, structure or functions of a protein usually have bad effects on the cell. Mistakes in splicing can also produce such molecules, so the inspection system – called nonsense-mediated mRNA decay (NMD) – needs to be on alert all the time.
NMD takes place when an RNA enters the cell cytoplasm, but scientists have connected it to splicing, which takes place before the RNA leaves the nucleus (Sun et al., 2000). The cell attaches a cluster of proteins to sites where introns have been removed, like attaching a sign on a boxcar to show that some of the following cars are missing. The cluster consists of at least six proteins and is called the exon junction complex (EJC). This complex is placed at splice sites and its position has an important influence on the fate of the RNA.
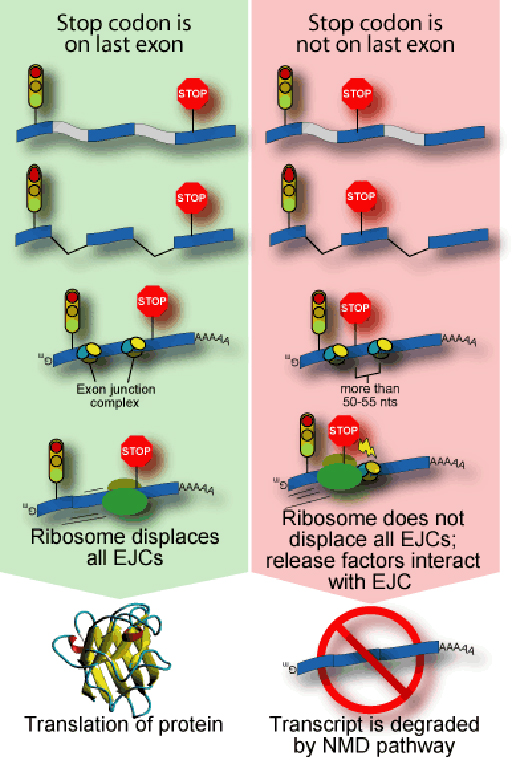
The translation of an RNA into protein is carried out by a molecular machine called the ribosome which docks onto a messenger RNA, reads its code and assembles a chain of amino acids that matches the sequence. Any EJCs on the molecule are simply moved out of the way. At the end of the protein-encoding part of the RNA, the ribosome encounters a three-letter signal called the stop codon and releases the finished protein. Mutations often alter the spelling of an RNA so that a stop codon appears somewhere in the middle of the molecule. This creates a code within the RNA that doesn’t make sense to the cell (the ‘nonsense’ in NMD) and may leave an EJC on the RNA, which promotes NMD (see figure).
To help prevent potentially harmful proteins from being produced, NMD recognises EJCs that are found more than about 50 nucleotides after a stop codon. The process of translation is interrupted, and other molecules come to carry the RNA away and break it down.
But some RNAs escape NMD and produce harmful proteins which can lead to disease. Even when NMD works, the result may be disease, because NMD may remove an RNA that is damaged but nevertheless necessary. In 1989, Lynne Maquat’s laboratory at the Roswell Park Memorial Institute (New York, USA) showed that NMD contributes to beta-thalassaemia, the most common genetic disease in the Western world. Beta-thalassaemia reduces the body’s production of haemoglobin, which is needed to carry oxygen through the blood. The disease arises in people who inherit a mutant form of a gene called beta-globin; NMD catches the mutation and the body breaks down beta-globin RNA – removing an important molecule (Lim et al., 1989). In this case, an intended safety mechanism is actually attacking the body.
Until recently, NMD was considered to be little more than a means of trapping RNAs with errors; now it is known to be a more general tool that the cell uses to control the quantity and quality of certain molecules. This happens because the normal process of alternative splicing sometimes produces RNAs with nonsense codons; for some reason, the cut-and-paste operation produces a bit of nonsense code in the middle of an RNA. In 2004, R. Tyler Hillman, Richard Green and Steven Brenner of the University of California, Berkeley, USA, carried out a computer analysis which showed that about one-third of the time, alternative splicing places a stop codon more than 50 nucleotides in front of a splice site. This activates NMD, which eliminates most of the RNA before it can be transformed into proteins (Hillman et al., 2004).
The same year, Harry Dietz’s group at John Hopkins University School of Medicine (Maryland, USA) studied this effect in mammalian cells. They shut down the NMD machinery by removing a protein called Upf1, which is essential for the process. This changed the behaviour of a huge number of genes: about 10% of the genes they studied became more productive, probably because spliced forms were slipping through that normally would have been caught by NMD and destroyed (Mendell et al., 2004).
This means that NMD, like alternative splicing, is a commonly used side-street on the road between genes and proteins. There are many more, some of which will be discussed in a future issue of Science in School. If the suspense is too great, here is a riddle to keep you busy: the colour of purple petunias is due to a single copy of a particular gene. What happens to the petunia’s colour if you add a second copy of that gene to the flower?
References
- Berget SM, Moore C, Sharp PA (2000). Spliced segments at the 5’ terminus of adenovirus 2 late mRNA. Reviews in Medical Virology 10: 355-371
- Castillo-Davis CI, Mekhedov SL, Hartl DL, Koonin EV, Kondrashov FA (2002) Selection for short introns in highly expressed genes. Nature Genetics 31: 415-418. doi:10.1038/ng940
- Crick F (1970) Central Dogma of Molecular Biology. Nature 227: 561-563
- Download the article free of charge here [3.3 MB], or subscribe to Nature today: www.nature.com/subscribe
- Hillman RT, Green RE, Brenner SE (2004) An unappreciated role for RNA surveillance. Genome Biology 5: R8. doi:10.1186/gb-2004-5-2-r8
- Lim S, Mullins JJ, Chen CM, Gross KW, Maquat LE (1989) Novel metabolism of several beta zero-thalassemic beta-globin mRNAs in the erythroid tissues of transgenic mice. The EMBO Journal 8(9): 2613-2619
- Lipscombe D (2005) Neuronal proteins custom designed by alternative splicing. Current Opinion in Neurobiology 15: 358-363. doi:10.1016/j.conb.2005.04.002
- Losson R, Lacroute F (1979) Interference of nonsense mutations with eukaryotic messenger RNA stability. Proceedings of the National Academy of Sciences USA 76: 5134-5137
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC (2004) Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nature Genetics 36: 1073-1078. doi:10.1038/ng1429
- Raible F et al. (2005) Vertebrate-type intron-rich genes in the marine annelid. Platynereis dumerilii. Science 310: 1325-1326. doi:10.1126/science.1119089
- Sun X, Moriarty PM, Maquat LE (2000) Nonsense-mediated decay of glutathione peroxidase 1 mRNA in the cytoplasm depends on intron position. EMBO Journal 19: 4734-4744. doi:10.1093/emboj/19.17.4734
Web References
- w1 – The transcript of Philip Sharp’s Nobel lecture about alternative splicing, ‘Split Genes and RNA Splicing’, is available online.
- A brief overview of Richard Roberts and Philip Sharp’s work is given in the press release announcing their Nobel Prize.
- For more information about the Nobel Prize, including biographies of the prize-winners, see here.
Resources
- Many papers by Crick are freely available on the Nature website.
Institutions
Review
Advances in biotechnology over the last 40 years have impacted on areas such as agriculture, food science and medicine. Turning our scientific knowledge of the central dogma of molecular biology into useful applications, especially in research into disease and medical treatments, requires a more detailed understanding of the functions of the molecules of life.
This series will introduce advances and new theories which may be too recent to appear in textbooks. It will also illustrate the international and universal quest to learn more about how the molecules of life operate.
This article provides excellent background reading and parts of it could be used to increase and test students’ understanding of the central dogma, RNA properties or protein synthesis. In particular, the article would be applicable to considerations of the uses of biotechnology, the importance of ongoing research and the recognition of scientific endeavour.
Marie Walsh, Republic of Ireland





