Outmanoeuvering influenza’s tricks Understand article
Catching the influenza virus can be more than just a nuisance: these pathogens have caused the most deadly pandemic in recent history. Claire Ainsworth investigates how scientists are working to prevent it happening again.
Image courtesy of EMBL Photolab
It seems like a constant battle – no sooner have you downloaded a security upgrade for your computer than a hacker somewhere finds another weak point. Once again, your machine is open to attack by viruses, spyware and other malicious programs, until the next upgrade patches the flaw.
The cells in our bodies face similar problems: they too have a multitude of security systems to prevent their biochemistry from being hijacked by viruses. But these systems are not perfect, and many viruses have evolved sneaky strategies to hack into them. A group of scientists from the FLUPOLw1 EU initiative are trying to understand these strategies to develop a security upgrade for the human body. They have laid bare one of the tactics used by the influenza virus, paving the way for new drugs to combat future flu pandemics.
Influenza is a grave concern for governments and health organisations around the world. Although common human strains of the virus are rarely deadly to healthy people, they can prove fatal to the elderly or to those with other illnesses or weakened immune systems. Worse, new strains of the virus are constantly evolving and could prove to be more virulent than their predecessors. The most worrying threat of all is the potential for bird flu – the influenza virus that infects birds – to develop the ability to infect humans easily. In the past, viruses that have made this leap have been extraordinarily deadly. For example, the 1918 influenza pandemic (from Greek pan: all, and demos: people, an epidemic of infectious disease that spreads through populations across a large region, such as a continent, or even worldwide), also originated in birds and killed between 50 and 100 million people as it swept around the world. It was the deadliest pandemic in recent history.
In the past few years, fears have been mounting over a strain of bird flu called H5N1 (see also Niekoop & Rienks, 2006), which has already infected and killed a few hundred people worldwide. Should it eventually acquire the ability to spread from human to human, we could face a new, deadly pandemic. Flu vaccines do exist, but those that work against one flu strain often have a limited effect against others. Vaccines against new strains take a long time to produce, and so may come too late to curtail a new pandemic. New ways to halt the spread of the virus are badly needed.
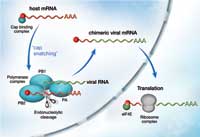
steals the cap of host mRNA
with the help of protein PB2,
which makes part of the
polymerase complex. With the
cap attached, the viral RNA can
be translated into proteins by
ribosomes in the cytoplasm and
more copies of the virus are
produced. Click to enlarge image
Image courtesy of Petra Riedinger
When the influenza virus infects a host cell, its goal is to produce many copies of itself that go on to attack even more cells. A viral enzyme composed of three subunits, called polymerase, is the key to this process. It copies the genetic material of the virus and steers the host cell machinery towards the synthesis of viral proteins. It does this by a process known as ‘cap snatching’ (see image). The cap is a short extra piece of RNA, which must be present at the beginning of all messenger RNAs (mRNAs). It is required to direct the cell’s protein-synthesis machinery to its starting point, so that it can synthesise a new protein according to the information laid down in the mRNA. The viral polymerase steals the cap from host-cell RNA molecules: it binds to host-cell mRNA via its cap, cuts the cap off and adds it to the beginning of the virus’ own mRNA. But exactly how the polymerase achieves this, and which of the three subunits does what, has remained controversial. The biochemistry of the viral polymerase is probably very different from human proteins, which makes it an attractive target for drugs aimed at reducing the spread of infection in the body, since this difference lowers the risk of damaging human proteins as a side effect.
Moreover, learning about the polymerase is crucial for understanding how influenza leaps between species. Several of the mutations that enable bird flu to adapt to life in a human host occur within its polymerase protein, so it will be essential to find out what they do. This is no easy task, and demands a wide range of expertise. The FLUPOL EU initiative was formed to meet this challenge: Stephen Cusack’s and Darren Hart’s groups at the EMBL outstation in Grenoblew2, France, together with colleagues from the Grenoble Unit of Virus Host Cell Interactionsw3, have joined forces with other European teams in Madrid, Lyon, London and Marburg. “We have a multidisciplinary approach to studying this enzyme, from bioinformatics via structure to mouse models of interspecies transmission,” says Stephen.
The polymerase, however, does not yield its secrets easily. For many years, it was impossible to get any detailed information on its three-dimensional structure. To do this, researchers need to produce the polymerase’s constituent proteins in a soluble form and get them to form crystals. By shining X-rays on the crystals, they are able to deduce the structures of these proteins and better understand how they work. For an introduction to crystallography, see the article by Dominique Cornuéjols in this issue. You can also try your hand at crystallising proteins in the classroom (Blattmann & Sticher 2009).
For a long time, no-one managed to get the all-important crystals. Whenever they tried, researchers found that the proteins either formed useless, insoluble lumps, or that they could not produce enough soluble protein to work on. Frustratingly, the scientists were unable to resort to their usual strategy for solving this problem: chopping the proteins up into their individual working parts, called domains, in the hope that these might be soluble. This method relies on being able to identify the domains in advance. Biologists usually do so by comparing the amino-acid sequence of the target protein with those of other proteins whose structures are known. But the flu polymerase is unlike any other known protein, so such an approach was impossible.

Image courtesy of EMBL Photolab
The solution came in the form of a new automated system Darren invented. Called ESPRIT (Expression of Soluble Proteins by Random Incremental Truncation), this system allows scientists to screen all the possible fragments of a protein for suitable soluble domains quickly and easily. In 2007, Darren and Stephen used ESPRIT to obtain the first soluble fragment of PB2, one of the three subunits (PA, PB1, PB2) that combine to form influenza’s polymerase (Tarendeau et al., 2007). Having determined the structure of this fragment, they discovered how it helps the PB2 subunit hitchhike into the cell’s nucleus, where it assembles with the other two subunits to form the functional polymerase.
Stephen and Darren were particularly interested in another region of PB2 which plays a key role in cap snatching. The PB2 subunit binds to host-cell mRNAs via the cap, and then the polymerase’s PB1 subunit cuts the cap off and adds it to the beginning of the viral mRNA. The capped viral mRNA can then be recognised by the host-cell machinery, allowing viral proteins to be made at the expense of host-cell proteins. Without a cap, an mRNA cannot be turned into protein. So drugs that block PB2’s cap-binding activity could stop the flu virus reproducing.
Until now, however, little was known about how PB2 worked. “This has been studied for many years, but no one was sure exactly where the cap binding domain was,” says Stephen.
Thanks to ESPRIT, Darren, Stephen and colleagues found a PB2 fragment that bound to the cap. With the help of the high-throughput crystallisation facility at EMBL Grenoble, and the intense synchrotron X-ray beamlines at the European Synchrotron Radiation Facility (ESRF)w4 (see Cornuéjols, 2009), they determined the structure of the PB2 cap-binding domain when bound to the cap. This revealed that the domain contained a structure never seen before. “It’s completely unlike any other protein in its fold,” says Stephen.
But although the structure was entirely new, the way the domain interacted with the cap uses the same basic idea as other cap-binding proteins: the central interaction is a ‘sandwich’ with two PB2 amino acids stacking either side of the cap (see image). “It’s a nice example of convergent evolution, where proteins from completely different origins come up with the same solution,” says Darren.
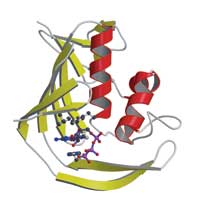
the influenza virus’ PB2 protein
shows how the virus steals a ‘cap’
molecule from its host. PB2 binds
the cap (labelled) by sandwiching
it between aromatic amino acids.
Click to enlarge image
Image courtesy of Petra Riedinger
To prove that their fragment was indeed the culprit cap-stealer, Darren and Stephen created fragments containing mutations of the amino acids which sandwich the cap. Sure enough, most of the fragments with mutations didn’t interact with the cap. What’s more, as shown by FLUPOL collaborators at the Centro Nacional de Biotecnologia in Madridw5, the same mutations blocked the ability of the whole polymerase to make new viruses. “We were able to display the critical importance of this for the virus as a whole,” says Stephen (Guilligay et al., 2008).
Now that the structure is known, scientists can use it to design relatively simple, small molecules that block PB2’s action. “If you can inhibit cap-snatching, you can kill the virus,” says Stephen. He and Darren are now gearing up to look for potential drugs that do just that. They are using the structure to actively design an inhibitor, but they are also exploiting their ability to produce large amounts of the PB2 fragment to search vast chemical compound collections for other molecules PB2 may bind to – potential PB2 inhibitors. “We are the first people to be able to make enough of this fragment to take this brute-force approach for identifying small molecule inhibitors,” says Darren.
Recently, researchers at the UVHCI and in Stephen’s group discovered that part of the PA subunit, the third viral polymerase’s subunit, is responsible for cleaving the cap off the host mRNA (Dias et al., 2009). The results came as a big surprise, because everybody thought that the cleaving activity resides in a different part of the polymerase. So with PA, another promising antiviral drug target has been found.
The researchers produced crystals of the crucial PA domain and examined them with the powerful X-ray beams of the ESRF in Grenoble. The high-resolution images they obtained reveal the individual amino acids that constitute the active site responsible for separating the cap from the RNA. A hollow canyon in the centre of the PA domain captures the long mRNA strand, and the metal complexes at the top edges of the canyon cut off the cap. Further experiments at ESRF confirmed that these metal complexes contain manganese, providing an important hint for drug development, since it will be essential to take the metal ions into account for designing improved inhibitors. The trick will be to exploit the knowledge about the structure of the viral polymerase subunit’s active sites – which either are responsible for ligand binding or have enzymatic activity – to design small molecule inhibitors which can specifically block the sites and therefore virus reproduction.
Their findings have given scientists a close-to-complete picture of the cap-snatching mechanism, and the researchers are now in the process of using ESPRIT to crack the rest of the flu polymerase’s structure in the hope that, in the near future, they will uncover new ways of countering influenza’s dirty tricks.
References
- Blattmann B, Sticher P (2009) Growing crystals from protein. Science in School 11: 30-36.
- Cornuéjols D (2009) Biological crystals: at the interface between physics, chemistry and biology. Science in School 11: 70-76.
- Dias A, Bouvier D, Crépin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, Ruigrok RW (2009) The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458: 914-918. doi: 10.1038/nature07745. Download the article free of charge here or subscribe to Nature today: www.nature.com/subscribe
- Guilligay D Tarendeau F, Resa-Infante P, Coloma R, Crepin T, Sehr P, Lewis J, Ruigrok RW, Ortin J, Hart DJ, Cusack S (2008) The structural basis for cap-binding by influenza virus polymerase subunit PB2. Nature Structural and Molecular Biology 15(5): 500-106. doi: 10.1038/nsmb.1421
- Niekoop L, Rienks F (2006) The ecologist’s view of bird flu. Science in School 3: 24-30.
- Tarendeau F, Boudet J, Guilligay D, Mas PJ, Bougault CM, Boulo S, Baudin F, Ruigrok RW, Daigle N, Ellenberg J, Cusack S, Simorre JP, Hart DJ (2007) Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nature Structural and Molecular Biology 14(3): 229-33. doi: 10.1038/nsmb1212
Web References
- w1 – For more information on the EU FLUPOL initiative, see: www.flupol.eu
- w2 – You can find the website of the European Molecular Biology Laboratory (EMBL) outstation in Grenoble here: www.embl.fr
- w3 – To find out more about the Unit of Virus Host Cell Interactions in Grenoble, see: www.uvhci.fr
- w4 – For more information on the European Synchrotron Radiation Facility (ESRF) in Grenoble, see: www.esrf.eu
- w5 – To learn more about the Centro Nacional de Biotecnologia in Madrid, see: www.cnb.uam.es





