Uracil in DNA: error or signal? Understand article
Uracil is well known as one of the bases used in RNA, but why is it not used in DNA – or is it? Angéla Békési and Beáta G Vértessy investigate.
Thymine versus uracil
Our genetic information is stored in the form of DNA, using a four-letter alphabet. The four ‘letters’ correspond to the four chemical bases that each building block of DNA – called a nucleotide – can have: adenine (A), thymine (T), cytosine (C) and guanine (G; see Figure 1). As James Watson and Francis Crick famously discovered, DNA forms a double helix in which the four bases always pair up the same way, through specific hydrogen bonds: adenine binds to thymine, and guanine to cytosine (see Figures 2 and 3).
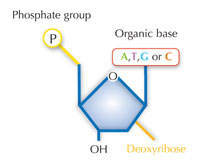
Image courtesy of Nicola Graf
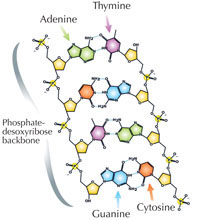
Image courtesy of Madeleine Price Ball; image source: Wikimedia Commons
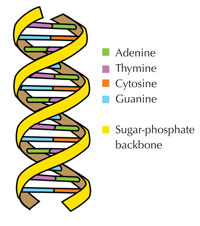
Image courtesy of Forluvoft; image source: Wikimedia Commons
There is an alternative fifth letter, though: uracil (U), which forms the same pattern of hydrogen bonds with adenine (see Figure 4). But although uracil is commonly used in RNA, this is not the case in DNA, where thymine is used instead. Why might this be?
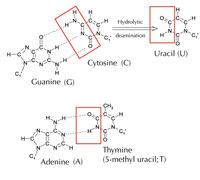
Image courtesy of Angéla Békési
Chemically, thymine is a uracil molecule with an extra methyl group attached. What would be the advantage, in evolutionary terms, of using this more complex building block in DNA? The answer may lie in how cells correct damage to DNA.
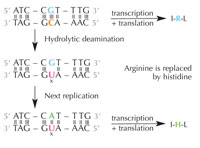
deamination of cytosine can
change the amino acids
encoded by the sequence.
Click to enlarge image
Image courtesy of Nicola Graf
Cytosine can spontaneously turn into uracil, through a process called hydrolytic deamination (see Figure 4). When this happens, the guanine that was initially bound to that cytosine molecule is left opposite uracil instead (remember that uracil normally binds to adenine). When the cell next replicates its DNA, the position opposite this uracil molecule would be taken up by an adenine instead of the guanine that should be there, altering the message that this section of DNA encodes (see Figure 5). This process of cytosine deamination is one of the most common types of DNA damage, but is normally corrected effectively. How does the cell do this?
deamination.
Click to enlarge image
Image courtesy of Nicola Graf
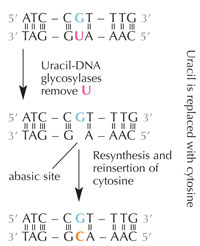
Cells have a repair system that can detect when a uracil is sitting where a cytosine should be, and correct the mistake before it is replicated and passed on. The complex machinery to do that consists of several enzymes: first uracil-DNA glycosylases recognise the uracil, and cut it out of the DNA. Then several enzymes contribute to the elimination and re-synthesis of the damaged part of DNA, during which the abasic (‘empty’) site in the DNA is replaced with a cytosine (see Figure 6).
However, the most common form of uracil-DNA glycosylase cannot tell which base the uracil is paired with, i.e. whether the uracil was intended to be there (if bound with adenine) or if it is a mutated cytosine (and is opposite guanine); instead, it would recognise and cut out both types of uracil. Clearly, this would cause problems. The solution to this potential problem is thought to have been the evolution of a mechanism in which ‘correct’ uracils (paired with adenine) were labelled with a methyl group – resulting in thymine. This way, if the cell machinery found a uracil, it cut it out and repaired it, but if it found a uracil with a methyl label – a thymine (see Figure 4) – it left it. Over time, therefore, thymine in DNA became the standard instead of uracil, and most cells now use uracil only in RNA.
Why was uracil retained in RNA? RNA is more short-lived than DNA and – with a few exceptions – is not the repository for long-term storage of genetic information, so cytosine molecules that spontaneously turn into uracils in RNA do not present a great threat to the cell. Thus, there was probably no evolutionary pressure to replace uracil with the more complex (and presumably more costly) thymine in RNA.
Thymine-less cell death
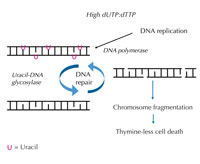
increases, DNA polymerase
frequently incorporates uracil
instead of thymine during
both replication and repair.
Uracil-DNA glycosylase
removes the uracil and
initiates further repair
involving DNA strand breaks
in an intermediate step.
Repair synthesis, however,
may reintroduce uracil,
leading to a futile DNA repair
cycle. Eventually, the system
is overloaded and
chromosome fragmentation
occurs, leading to cell death.
Click to enlarge image
Image courtesy of Angéla
Békési
When DNA is synthesised, the DNA polymerase enzymes (which catalyse the synthesis) cannot discriminate between thymine and uracil. They only check whether the hydrogen bonds form correctly, i.e. whether the base pairs are matched properly. To these enzymes, it does not matter whether thymine or uracil binds to adenine. Normally, the amounts of deoxyuridine triphosphate (dUTP, a source of uracil) in the cell are kept very low compared to levels of deoxythymidine triphosphate (dTTP, a thymine source), preventing uracil incorporation during DNA synthesis.
If this strict regulation is perturbed and the ratio of dUTP to dTTP rises, the amount of uracil that is incorrectly incorporated into DNA also increases. The repair system – which, unlike DNA polymerases, can distinguish uracil from thymine – then attempts to cut out the uracil with the help of uracil-DNA glycosylase and to re-synthesise the DNA, which involves temporarily cleaving (cutting) the DNA backbone. However, if the ratio of dUTP to dTTP is still elevated, this re-synthesis may again incorporate uracil instead of thymine. This cycle eventually leads to DNA strand breaks and chromosome fragmentation, when these temporary cuts in the DNA happen one after the other and too close to each other (see Figure 7). This results in a specific type of programmed cell death, called thymine-less cell death.
The process of thymine-less cell death can be deliberately exploited in the treatment of cancer. Because cancer cells proliferate at such a high rate compared to normal cells, they synthesise a greater amount of DNA per given time period and therefore require large amounts of dUTP. By raising the ratio of dUTP to dTTP, these cancer cells can be selectively targeted and eliminated.
Uracil DNA still exists
Although most cells use uracil for RNA and thymine for DNA, there are exceptions. Some organisms have uracil instead of thymine in all their DNA, and other organisms have uracil in only some of their DNA. What could be the evolutionary advantage of that? Let’s take a look at some examples.
Uracil in viral DNA
phage virus infecting a
bacterial cell
Image courtesy of cdascher /
iStockphoto
Two species of phage (viruses that infect bacteria) are known to have DNA genomes with only uracil and no thymine. We do not yet know whether these phages are representatives of an ancient life form that never evolved thymine DNA, or whether their uracil-substituted genomes are a newly evolved strategy. Nor do we know why these phages use uracil instead of thymine, but it may play an essential role in the life cycle of these viruses. If that is the case, it would make sense for the viruses to ensure that the uracil in their DNA is not replaced with thymine. And one of these phages has in fact been shown to have a gene that encodes a specific protein to inhibit the host’s uracil-DNA glycosylase, thus preventing the viral genome from having its uracil ‘repaired’ by the host enzymes.
Programmed cell death in insect life cycles

lack the enzyme capable of
removing uracil from their
DNA
Image courtesy of spxChrome
/ iStockphoto and Nicola Graf
Uracil-DNA also appears to play a role in the development of endopterygotes – insects that undergo pupation during their life cycle (ants and butterflies do; grasshoppers and termites do not). These insects lack the main gene for uracil-DNA glycosylase, which would otherwise remove uracil from their DNA.
Moreover, our own research has shown that, in larvae of the fruit fly Drosophila melanogaster, the ratio of dUTP to dTTP is regulated in an unusual manner: in all tissues that will not be needed in the adult insect, there are much lower levels of the enzyme that breaks down dUTP and generates a precursor for dTTP production. Consequently, significant amounts of uracil are incorporated into these tissues during DNA synthesis.
So during the larval stages, uracil-DNA is produced and seems not to be corrected in tissues that are to be degraded during the pupal stage. As these insects lack the main uracil-DNA glycosylase enzyme, at the pupal stage, additional uracil-DNA-specific factors may recognise this accumulated uracil as a signal to initiate cell death. We have already identified an insect-specific protein that seems to be capable of degrading uracil-DNA, and we are investigating whether this enzyme is used to initiate programmed cell death.

antibody gene sequences
elicits a DNA repair response,
which has the effect of
increasing antibody protein
diversity. An extensive
antibody pool increases the
chance of the immune
system recognising
unwanted invaders
Image courtesy of taramol /
iStockphoto
Beneficial errors: the vertebrate immune system
Uracil in DNA, however, can also be found closer to home – in the immune system of vertebrates like us. Part of our immune system, the adaptive immune system, produces a large number of different antibodies that are trained to protect us from specific pathogens. To increase the number of different antibodies that can be created, we shuffle the DNA sequence in the regions that code for them, not only by recombining the existing sequences in the cells but also by creating new ones through vastly increased mutation rates, known as hypermutation.
Hypermutation starts with a specific enzyme (an activation-induced deaminase) that changes cytosine into uracil (see Figure 4) at specific DNA loci, eliciting an error-prone repair response, which the organism uses to its advantage: ‘errors’ generate new sequences that can be used to make different antibodies. This system is very strictly regulated, however, as if it got out of hand, it would lead to cancer.
When considering the question of why uracil or why thymine, we need to consider the evolutionary context. Living organisms have evolved in a continuously changing environment, facing a dynamic set of challenges. Thus, a solution that avoids mistakes being incorporated into DNA is advantageous to most organisms and most cells, which explains why thymine-DNA became the norm. Under certain circumstances, however, ‘mistakes’ themselves can be beneficial, which is why some cells still use uracil in their DNA.
Resources
- To find out more about the work of Beáta Vértessy’s research group, see: www.enzim.hu/~vertessy
- To download a summary of Villő Muha’s PhD thesis, which was written under Beáta Vértessy’s supervision and focuses on uracil-DNA in Drosophila melanogaster, use the link: http://teo.elte.hu/minosites/tezis2010_angol/v_muha.pdf
- The complete thesis is available here: http://teo.elte.hu/minosites/ertekezes2010/muha_v.pdf
Review
This article demonstrates that science never sleeps, shaking up the dogma that uracil only exists in RNA. As the article explains, this is not always the case. And even when it is, why should that be?
To help the students understand the article, guiding questions could be:
- Describe the bonding structures between the two complementary base pairs in DNA.
- Which of the bases is replaced in RNA?
- Describe and draw a graph of the repair enzyme pathway triggered when uracil is found in DNA.
- The ratio of which molecules could be adjusted to stop cancerous cells from growing and dividing?
- Why is uracil ‘tolerated’ in RNA?
- Which living organisms use uracil DNA and how?
Friedlinde Krotscheck, Austria





